This is a fixed-text formatted version of a Jupyter notebook.
You can contribute with your own notebooks in this GitHub repository.
Source files: image_fitting_with_sherpa.ipynb | image_fitting_with_sherpa.py
CTA 2D source fitting with Sherpa¶
- Acero & Y. Gallant October 2017
Introduction¶
Sherpa is the X-ray satellite Chandra modeling and fitting application. It enables the user to construct complex models from simple definitions and fit those models to data, using a variety of statistics and optimization methods. The issues of constraining the source position and morphology are common in X- and Gamma-ray astronomy. This notebook will show you how to apply Sherpa to CTA data.
Here we will set up Sherpa to fit the counts map and loading the ancillary images for subsequent use. A relevant test statistic for data with Poisson fluctuations is the one proposed by Cash (1979). The simplex (or Nelder-Mead) fitting algorithm is a good compromise between efficiency and robustness. The source fit is best performed in pixel coordinates.
Read sky images¶
The sky image that are loaded here have been prepared in a separated notebook. Here we start from those fits file and focus on the source fitting aspect.
The info needed for sherpa are: - Count map - Background map - Exposure map - PSF map
For info, the fits file are written in the following way in the Sky map generation notebook:
images['counts'] .write("G300-0_test_counts.fits", clobber=True)
images['exposure'] .write("G300-0_test_exposure.fits", clobber=True)
images['background'].write("G300-0_test_background.fits", clobber=True)
##As psf is an array of quantities we cannot use the images['psf'].write() function
##all the other arrays do not have quantities.
fits.writeto("G300-0_test_psf.fits",images['psf'].data.value,overwrite=True)
In [1]:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from gammapy.image import SkyImage
from astropy.coordinates import SkyCoord
import astropy.units as u
from astropy.io import fits
from astropy.wcs import WCS
# You may see a Warning concerning XSPEC
# As we will note use Xspec spectral models this warning is not important
import sherpa.astro.ui as sh
WARNING: imaging routines will not be available,
failed to import sherpa.image.ds9_backend due to
'RuntimeErr: DS9Win unusable: Could not find xpaget on your PATH'
WARNING: failed to import WCS module; WCS routines will not be available
WARNING: failed to import sherpa.astro.xspec; XSPEC models will not be available
In [2]:
# Read the fits file to load them in a sherpa model
hdr = fits.getheader("G300-0_test_counts.fits")
wcs = WCS(hdr)
sh.set_stat("cash")
sh.set_method("simplex")
sh.load_image("G300-0_test_counts.fits")
sh.set_coord("logical")
sh.load_table_model("expo", "G300-0_test_exposure.fits")
sh.load_table_model("bkg", "G300-0_test_background.fits")
sh.load_psf("psf", "G300-0_test_psf.fits")
In principle one might first want to fit the background amplitude. However the background estimation method already yields the correct normalization, so we freeze the background amplitude to unity instead of adjusting it. The (smoothed) residuals from this background model are then computed and shown.
In [3]:
sh.set_full_model(bkg)
bkg.ampl = 1
sh.freeze(bkg)
data = sh.get_data_image().y - sh.get_model_image().y
resid = SkyImage(data=data, wcs=wcs)
resid_table = [] # Keep residual images in a list to show them later
resid_smo6 = resid.smooth(radius = 6)
resid_smo6.plot()
resid_table.append(resid_smo6)
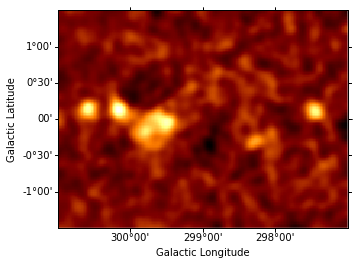
Find and fit the brightest source¶
We then find the position of the maximum in the (smoothed) residuals map, and fit a (symmetrical) Gaussian source with that initial position:
In [4]:
maxcoord = resid_smo6.lookup_max()
maxpix = resid_smo6.wcs_skycoord_to_pixel(maxcoord[0])
sh.set_full_model(bkg + psf(sh.gauss2d.g0) * expo) # creates g0 as a gauss2d instance
g0.xpos = maxpix[0]
g0.ypos = maxpix[1]
sh.freeze(g0.xpos, g0.ypos) # fix the position in the initial fitting step
expo.ampl = 1e-9 # fix exposure amplitude so that typical exposure is of order unity
sh.freeze(expo)
sh.thaw(g0.fwhm, g0.ampl) # in case frozen in a previous iteration
g0.fwhm = 10 # give some reasonable initial values
g0.ampl = maxcoord[1]
sh.fit() # Performs the fit; this takes a little time.
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47907.2
Final fit statistic = 47503 at function evaluation 234
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 404.211
g0.fwhm 6.43418
g0.ampl 0.400131
Fit all parameters of this Gaussian component, fix them and re-compute the residuals map.
In [5]:
sh.thaw(g0.xpos, g0.ypos)
sh.fit()
sh.freeze(g0)
data = sh.get_data_image().y - sh.get_model_image().y
resid = SkyImage(data=data, wcs=wcs)
resid_smo6 = resid.smooth(radius = 6)
resid_smo6.show(vmin = -0.5, vmax = 1)
resid_table.append(resid_smo6)
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47503
Final fit statistic = 47498.4 at function evaluation 354
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 4.57486
g0.fwhm 6.33581
g0.xpos 41.9218
g0.ypos 81.3048
g0.ampl 0.417545
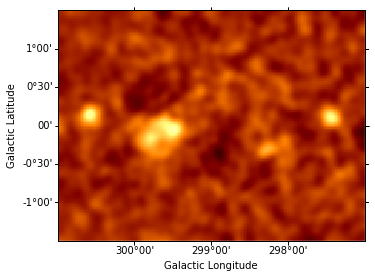
Iteratively find and fit additional sources¶
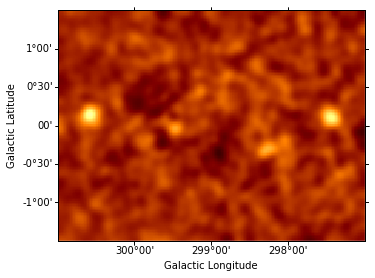
Instantiate additional Gaussian components, and use them to iteratively fit sources, repeating the steps performed above for component g0. (The residuals map is shown after each additional source included in the model.) This takes some time…
In [6]:
for i in range(1,6):
sh.create_model_component('gauss2d', 'g' + str(i))
gs = [g0, g1, g2, g3, g4, g5]
sh.set_full_model(bkg + psf(g0+g1+g2+g3+g4+g5) * expo)
for i in range(1, len(gs)) :
gs[i].ampl = 0 # initialize components with fixed, zero amplitude
sh.freeze(gs[i])
for i in range(1, len(gs)) :
maxcoord = resid_smo6.lookup_max()
maxpix = resid_smo6.wcs_skycoord_to_pixel(maxcoord[0])
gs[i].xpos = maxpix[0]
gs[i].ypos = maxpix[1]
gs[i].fwhm = 10
gs[i].fwhm = maxcoord[1]
sh.thaw(gs[i].fwhm)
sh.thaw(gs[i].ampl)
sh.fit()
sh.thaw(gs[i].xpos)
sh.thaw(gs[i].ypos)
sh.fit()
sh.freeze(gs[i])
data = sh.get_data_image().y - sh.get_model_image().y # estimate residual map = data - model
resid = SkyImage(data=data, wcs=wcs)
resid_smo6 = resid.smooth(radius = 6)
resid_smo6.show(vmin = -0.5, vmax = 1)
resid_table.append(resid_smo6)
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47498.4
Final fit statistic = 47335 at function evaluation 269
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 163.402
g1.fwhm 19.8613
g1.ampl 0.100529
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47335
Final fit statistic = 47293.7 at function evaluation 394
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 41.3431
g1.fwhm 19.9265
g1.xpos 67.2544
g1.ypos 69.3013
g1.ampl 0.116012
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47293.7
Final fit statistic = 47185.5 at function evaluation 264
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 108.124
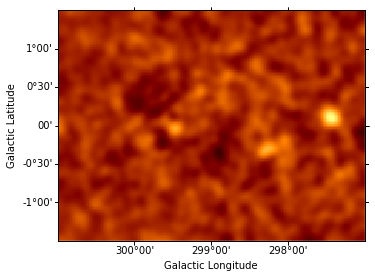
g2.fwhm 6.20212
g2.ampl 0.393345
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47185.5
Final fit statistic = 47182.5 at function evaluation 342
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 3.05173
g2.fwhm 6.19309
g2.xpos 20.8274
g2.ypos 81.6413
g2.ampl 0.40001
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47182.5
Final fit statistic = 47115 at function evaluation 275
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 67.4731
g3.fwhm 6.43218
g3.ampl 0.235481
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47115
Final fit statistic = 47114.3 at function evaluation 303
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 0.728928
g3.fwhm 6.38175
g3.xpos 177.458
g3.ypos 80.2233
g3.ampl 0.239637
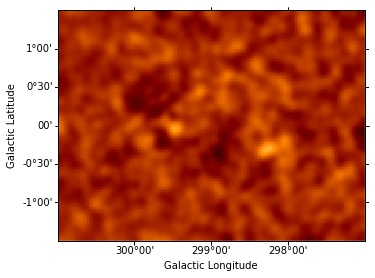
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47114.3
Final fit statistic = 47086.6 at function evaluation 277
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 27.7341
g4.fwhm 5.70151
g4.ampl 0.162191
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47086.6
Final fit statistic = 47085.7 at function evaluation 304
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 0.898776
g4.fwhm 5.94179
g4.xpos 135.942
g4.ypos 59.5875
g4.ampl 0.156213
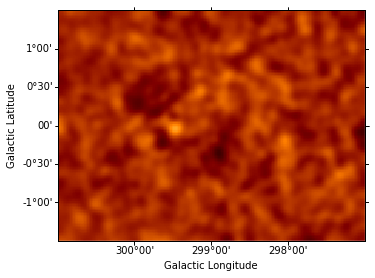
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47085.7
Final fit statistic = 47069 at function evaluation 262
Data points = 30000
Degrees of freedom = 29998
Change in statistic = 16.6424
g5.fwhm 2.82885
g5.ampl 0.52359
Dataset = 1
Method = neldermead
Statistic = cash
Initial fit statistic = 47069
Final fit statistic = 47068.9 at function evaluation 328
Data points = 30000
Degrees of freedom = 29996
Change in statistic = 0.0749129
g5.fwhm 2.82317
g5.xpos 76.1381
g5.ypos 73.1869
g5.ampl 0.524268
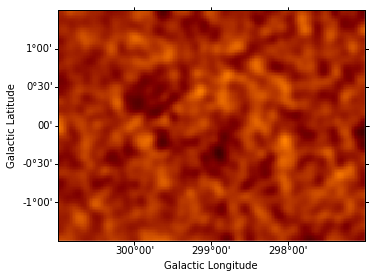
Generating output table and Test Statistics estimation¶
When adding a new source, one need to check the significance of this new source. A frequently used method is the Test Statistics (TS). This is done by comparing the change of statistics when the source is included compared to the null hypothesis (no source ; in practice here we fix the amplitude to zero).
\(TS = Cstat(source) - Cstat(no source)\)
The criterion for a significant source detection is typically that it should improve the test statistic by at least 25 or 30. The last excess fitted (g5) thus not a significant source:
In [7]:
from astropy.stats import gaussian_fwhm_to_sigma
from astropy.table import Table
rows = []
for idx, g in enumerate(gs):
ampl = g.ampl.val
g.ampl = 0
stati = sh.get_stat_info()[0].statval
g.ampl = ampl
statf = sh.get_stat_info()[0].statval
delstat = stati - statf
coord = resid.wcs_pixel_to_skycoord(g.xpos.val, g.ypos.val)
pix_scale = resid.wcs_pixel_scale()[0].deg
sigma = g.fwhm.val * pix_scale * gaussian_fwhm_to_sigma
rows.append(dict(
idx=idx,
delstat=delstat,
glon=coord.l.deg,
glat=coord.b.deg,
sigma=sigma ,
))
table = Table(rows=rows, names=rows[0])
table[table['delstat'] > 25]
Out[7]:
| idx | delstat | glon | glat | sigma |
|---|---|---|---|---|
| int64 | float64 | float64 | float64 | float64 |
| 0 | 136.582187364 | 300.151408614 | 0.136068174452 | 0.0538113987742 |
| 1 | 158.744570772 | 299.644884821 | -0.103966965279 | 0.169240266293 |
| 2 | 111.176173057 | 300.57305638 | 0.142771893381 | 0.0525992264273 |
| 3 | 68.2020558775 | 297.441233657 | 0.114422638212 | 0.0542015591058 |
| 4 | 28.632872396 | 298.27120691 | -0.298223927544 | 0.0504649169514 |
In [ ]:
# Small animation to show the source detection at each iteration
from ipywidgets.widgets.interaction import interact
def plot_resid(i):
fig, ax,cbar = resid_table[i].plot(vmin=-0.5, vmax=1,cmap='CMRmap')
# ax=plt.gca()
ax.set_title('CStat=%.2f'%(table['delstat'][i]))
ax.scatter(
table['glon'][i], table['glat'][i],
transform=ax.get_transform('galactic'),
color='none', edgecolor='azure', marker='o', s=400)
# plt.savefig('source_%i.png'%(i))
plt.show()
interact(plot_resid,i=(0,5))
Exercises¶
- If you look back to the original image: there’s one source that looks
like a shell-type supernova remnant.
- Try to fit is with a shell morphology model (use
sh.shell2d('shell')to create such a model). - Try to evaluate the
TSand probability of the shell model compared to a Gaussian model hypothesis - You could also try a disk model (use
sh.disk2d('disk')to create one)
- Try to fit is with a shell morphology model (use
What next?¶
These are good resources to learn more about Sherpa:
- https://python4astronomers.github.io/fitting/fitting.html
- https://github.com/DougBurke/sherpa-standalone-notebooks
You could read over the examples there, and try to apply a similar analysis to this dataset here to practice.
If you want a deeper understanding of how Sherpa works, then these proceedings are good introductions: