Fermi-LAT diffuse model predicted counts image¶
The SkyCube class allows for image-based analysis in energy
bands. In particular, similar functionality to gtmodel in the Fermi Science
tools [FSSC2013] is offered in compute_npred_cube
which generates a predicted instrument PSF-convolved counts cube based on a
provided background model. Unlike the science tools, this implementation is
appropriate for use with large regions of the sky.
Predicting Counts¶
The example script below computes the Fermi PSF-convolved predicted counts map
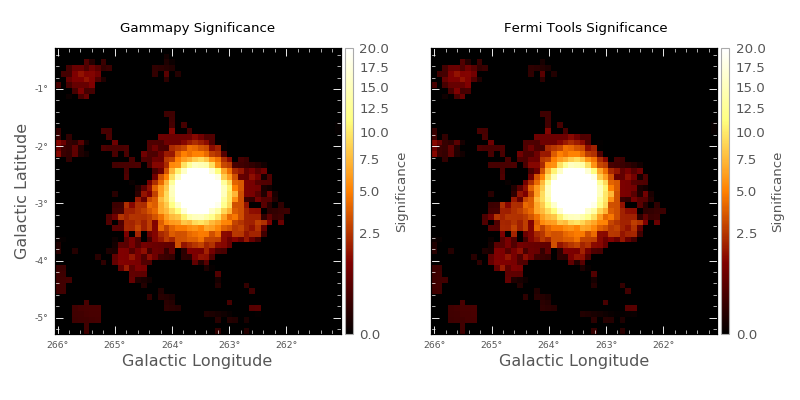
using SkyCube. This is then used to produce a Li & Ma significance
image [LiMa1983]. The left image shows the significance image,
while a comparison against the significance image
produced using the Fermi Science tools is shown on the right. These results are
for the Vela region for energies between 10 and 500 GeV.
"""Runs commands to produce convolved predicted counts map in current directory."""
import numpy as np
from astropy.coordinates import Angle
from astropy.units import Quantity
from astropy.io import fits
from astropy.wcs import WCS
from gammapy.cube import SkyCube, compute_npred_cube
from gammapy.datasets import FermiVelaRegion
from gammapy.irf import EnergyDependentTablePSF
from gammapy.utils.energy import EnergyBounds
__all__ = ['prepare_images']
def prepare_images():
# Read in data
fermi_vela = FermiVelaRegion()
background_file = FermiVelaRegion.filenames()['diffuse_model']
exposure_file = FermiVelaRegion.filenames()['exposure_cube']
counts_file = FermiVelaRegion.filenames()['counts_cube']
background_model = SkyCube.read(background_file, format='fermi-background')
exposure_cube = SkyCube.read(exposure_file, format='fermi-exposure')
# Re-project background cube
repro_bg_cube = background_model.reproject(exposure_cube)
# Define energy band required for output
energies = EnergyBounds([10, 500], 'GeV')
# Compute the predicted counts cube
npred_cube = compute_npred_cube(repro_bg_cube, exposure_cube, energies,
integral_resolution=5)
# Convolve with Energy-dependent Fermi LAT PSF
psf = EnergyDependentTablePSF.read(FermiVelaRegion.filenames()['psf'])
kernels = psf.kernels(npred_cube)
convolved_npred_cube = npred_cube.convolve(kernels, mode='reflect')
# Counts data
counts_cube = SkyCube.read(counts_file, format='fermi-counts')
counts_cube = counts_cube.reproject(npred_cube)
counts = counts_cube.data[0]
model = convolved_npred_cube.data[0]
# Load Fermi tools gtmodel background-only result
gtmodel = fits.open(FermiVelaRegion.filenames()['background_image'])[0].data.astype(float)
# Ratio for the two background images
ratio = np.nan_to_num(model / gtmodel)
# Header is required for plotting, so returned here
wcs = npred_cube.wcs
header = wcs.to_header()
return model, gtmodel, ratio, counts, header
"""Runs commands to produce convolved predicted counts map in current directory."""
import numpy as np
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.convolution import Tophat2DKernel
from scipy.ndimage import convolve
from gammapy.stats import significance
from aplpy import FITSFigure
from npred_general import prepare_images
model, gtmodel, ratio, counts, header = prepare_images()
# Top hat correlation
tophat = Tophat2DKernel(3)
tophat.normalize('peak')
correlated_gtmodel = convolve(gtmodel, tophat.array)
correlated_counts = convolve(counts, tophat.array)
correlated_model = convolve(model, tophat.array)
# Fermi significance
fermi_significance = np.nan_to_num(significance(correlated_counts, correlated_gtmodel,
method='lima'))
# Gammapy significance
significance = np.nan_to_num(significance(correlated_counts, correlated_model,
method='lima'))
titles = ['Gammapy Significance', 'Fermi Tools Significance']
# Plot
fig = plt.figure(figsize=(10, 5))
hdu1 = fits.ImageHDU(significance, header)
f1 = FITSFigure(hdu1, figure=fig, convention='wells', subplot=(1, 2, 1))
f1.set_tick_labels_font(size='x-small')
f1.tick_labels.set_xformat('ddd')
f1.tick_labels.set_yformat('ddd')
f1.show_colorscale(vmin=0, vmax=20, cmap='afmhot', stretch='sqrt')
f1.add_colorbar(axis_label_text='Significance')
f1.colorbar.set_width(0.1)
f1.colorbar.set_location('right')
hdu2 = fits.ImageHDU(fermi_significance, header)
f2 = FITSFigure(hdu2, figure=fig, convention='wells', subplot=(1, 2, 2))
f2.set_tick_labels_font(size='x-small')
f2.tick_labels.set_xformat('ddd')
f2.hide_ytick_labels()
f2.hide_yaxis_label()
f2.show_colorscale(vmin=0, vmax=20, cmap='afmhot', stretch='sqrt')
f2.add_colorbar(axis_label_text='Significance')
f2.colorbar.set_width(0.1)
f2.colorbar.set_location('right')
fig.text(0.15, 0.92, "Gammapy Significance")
fig.text(0.63, 0.92, "Fermi Tools Significance")
plt.tight_layout()
plt.show()
(Source code, png, hires.png, pdf)
Checks¶
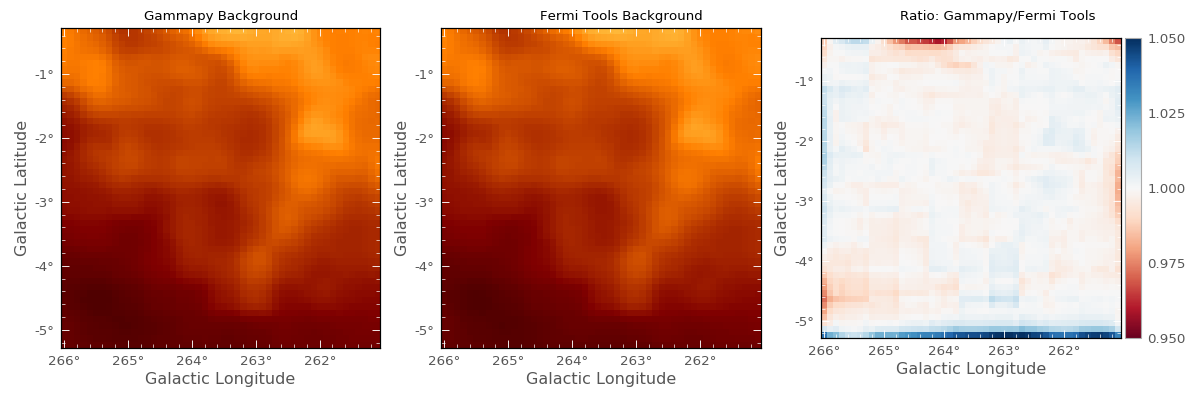
For small regions, the predicted counts cube and significance images may be checked against the gtmodel output. The Vela region shown above is taken in this example in one energy band with the parameters outlined in the README file for FermiVelaRegion.
Images for the predicted background counts in this region in the Gammapy case (left) and Fermi Science Tools gtmodel case (center) are shown below, based on the differential flux contribution of the Fermi diffuse model gll_iem_v05_rev1.fit. The image on the right shows the ratio. Note that the colorbar scale applies only to the ratio image.
"""Runs commands to produce convolved predicted counts map in current directory.
"""
import matplotlib.pyplot as plt
from astropy.io import fits
from aplpy import FITSFigure
from npred_general import prepare_images
model, gtmodel, ratio, counts, header = prepare_images()
# Plotting
fig = plt.figure(figsize=(15, 5))
image1 = fits.ImageHDU(data=model, header=header)
f1 = FITSFigure(image1, figure=fig, subplot=(1, 3, 1), convention='wells')
f1.show_colorscale(vmin=0, vmax=0.3, cmap='afmhot')
f1.tick_labels.set_xformat('ddd')
f1.tick_labels.set_yformat('dd')
image2 = fits.ImageHDU(data=gtmodel, header=header)
f2 = FITSFigure(image2, figure=fig, subplot=(1, 3, 2), convention='wells')
f2.show_colorscale(vmin=0, vmax=0.3, cmap='afmhot')
f2.tick_labels.set_xformat('ddd')
f2.tick_labels.set_yformat('dd')
image3 = fits.ImageHDU(data=ratio, header=header)
f3 = FITSFigure(image3, figure=fig, subplot=(1, 3, 3), convention='wells')
f3.show_colorscale(vmin=0.95, vmax=1.05, cmap='RdBu')
f3.tick_labels.set_xformat('ddd')
f3.tick_labels.set_yformat('dd')
f3.add_colorbar(ticks=[0.95, 0.975, 1, 1.025, 1.05])
fig.text(0.12, 0.95, "Gammapy Background")
fig.text(0.45, 0.95, "Fermi Tools Background")
fig.text(0.75, 0.95, "Ratio: Gammapy/Fermi Tools")
plt.tight_layout()
plt.show()
(Source code, png, hires.png, pdf)
We may compare these against the true counts observed by Fermi LAT in this region for the same parameters:
- True total counts: 1551
- Fermi Tools gtmodel predicted background counts: 265
- Gammapy predicted background counts: 282