Note
Click here to download the full example code or to run this example in your browser via Binder
Source detection and significance maps#
Build a list of significant excesses in a Fermi-LAT map.
Context#
The first task in a source catalogue production is to identify significant excesses in the data that can be associated to unknown sources and provide a preliminary parametrization in term of position, extent, and flux. In this notebook we will use Fermi-LAT data to illustrate how to detect candidate sources in counts images with known background.
Objective: build a list of significant excesses in a Fermi-LAT map
Proposed approach#
This notebook show how to do source detection with Gammapy using the
methods available in estimators. We will use images from a
Fermi-LAT 3FHL high-energy Galactic center dataset to do this:
perform adaptive smoothing on counts image
produce 2-dimensional test-statistics (TS)
run a peak finder to detect point-source candidates
compute Li & Ma significance images
estimate source candidates radius and excess counts
Note that what we do here is a quick-look analysis, the production of real source catalogs use more elaborate procedures.
We will work with the following functions and classes:
Setup#
As always, let’s get started with some setup …
import numpy as np
import astropy.units as u
# %matplotlib inline
import matplotlib.pyplot as plt
from IPython.display import display
from gammapy.datasets import MapDataset
from gammapy.estimators import ASmoothMapEstimator, TSMapEstimator
from gammapy.estimators.utils import find_peaks
from gammapy.irf import EDispKernelMap, PSFMap
from gammapy.maps import Map
from gammapy.modeling.models import PointSpatialModel, PowerLawSpectralModel, SkyModel
Check setup#
from gammapy.utils.check import check_tutorials_setup
check_tutorials_setup()
System:
python_executable : /Users/terrier/Code/anaconda3/envs/gammapy-dev/bin/python
python_version : 3.8.13
machine : x86_64
system : Darwin
Gammapy package:
version : 1.0rc2
path : /Users/terrier/Code/gammapy-dev/gammapy/gammapy
Other packages:
numpy : 1.22.4
scipy : 1.9.3
astropy : 5.1
regions : 0.6
click : 8.1.3
yaml : 6.0
IPython : 8.4.0
jupyterlab : 3.4.8
matplotlib : 3.5.3
pandas : 1.5.0
healpy : 1.16.1
iminuit : 2.17.0
sherpa : 4.15.0
naima : 0.10.0
emcee : 3.1.3
corner : 2.2.1
Gammapy environment variables:
GAMMAPY_DATA : /Users/terrier/Code/gammapy-dev/gammapy-data
Read in input images#
We first read the relevant maps:
counts = Map.read("$GAMMAPY_DATA/fermi-3fhl-gc/fermi-3fhl-gc-counts-cube.fits.gz")
background = Map.read(
"$GAMMAPY_DATA/fermi-3fhl-gc/fermi-3fhl-gc-background-cube.fits.gz"
)
exposure = Map.read("$GAMMAPY_DATA/fermi-3fhl-gc/fermi-3fhl-gc-exposure-cube.fits.gz")
psfmap = PSFMap.read(
"$GAMMAPY_DATA/fermi-3fhl-gc/fermi-3fhl-gc-psf-cube.fits.gz",
format="gtpsf",
)
edisp = EDispKernelMap.from_diagonal_response(
energy_axis=counts.geom.axes["energy"],
energy_axis_true=exposure.geom.axes["energy_true"],
)
dataset = MapDataset(
counts=counts,
background=background,
exposure=exposure,
psf=psfmap,
name="fermi-3fhl-gc",
edisp=edisp,
)
/Users/terrier/Code/anaconda3/envs/gammapy-dev/lib/python3.8/site-packages/astropy/wcs/wcs.py:725: FITSFixedWarning: 'datfix' made the change 'Set DATEREF to '2001-01-01T00:01:04.184' from MJDREF.
Set MJD-OBS to 54682.655283 from DATE-OBS.
Set MJD-END to 57236.967546 from DATE-END'.
warnings.warn(
Adaptive smoothing#
For visualisation purpose it can be nice to look at a smoothed counts image. This can be performed using the adaptive smoothing algorithm from Ebeling et al. (2006).
In the following example the ASmoothMapEstimator.threshold argument gives the minimum
significance expected, values below are clipped.
scales = u.Quantity(np.arange(0.05, 1, 0.05), unit="deg")
smooth = ASmoothMapEstimator(threshold=3, scales=scales, energy_edges=[10, 500] * u.GeV)
images = smooth.run(dataset)
plt.figure(figsize=(9, 5))
images["flux"].plot(add_cbar=True, stretch="asinh")
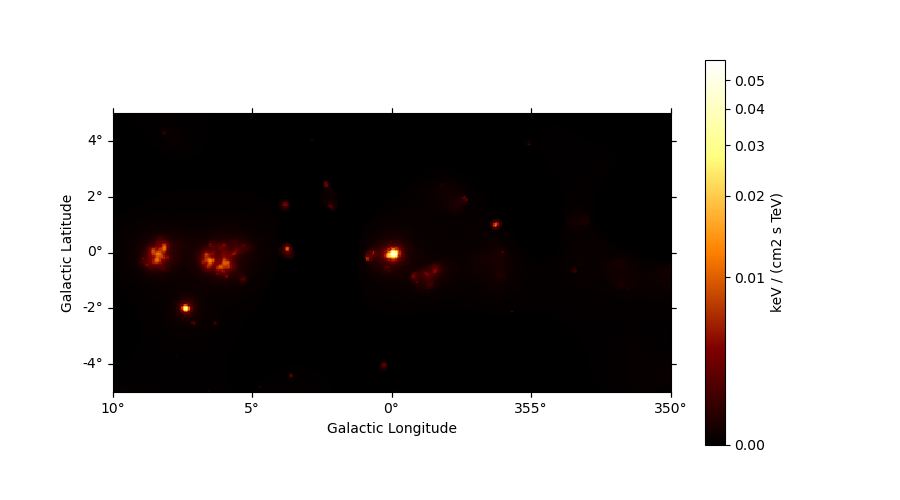
<WCSAxesSubplot:xlabel='Galactic Longitude', ylabel='Galactic Latitude'>
TS map estimation#
The Test Statistic, TS = 2 ∆ log L (Mattox et al. 1996), compares the likelihood function L optimized with and without a given source. The TS map is computed by fitting by a single amplitude parameter on each pixel as described in Appendix A of Stewart (2009). The fit is simplified by finding roots of the derivative of the fit statistics (default settings use Brent’s method).
We first need to define the model that will be used to test for the existence of a source. Here, we use a point source.
spatial_model = PointSpatialModel()
# We choose units consistent with the map units here...
spectral_model = PowerLawSpectralModel(amplitude="1e-22 cm-2 s-1 keV-1", index=2)
model = SkyModel(spatial_model=spatial_model, spectral_model=spectral_model)
estimator = TSMapEstimator(
model,
kernel_width="1 deg",
energy_edges=[10, 500] * u.GeV,
)
maps = estimator.run(dataset)
Plot resulting images#
fig, (ax1, ax2, ax3) = plt.subplots(
ncols=3,
figsize=(15, 3),
subplot_kw={"projection": counts.geom.wcs},
gridspec_kw={"left": 0.1, "right": 0.98},
)
maps["sqrt_ts"].plot(ax=ax1, add_cbar=True)
ax1.set_title("Significance map")
maps["flux"].plot(ax=ax2, add_cbar=True, stretch="sqrt", vmin=0)
ax2.set_title("Flux map")
maps["niter"].plot(ax=ax3, add_cbar=True)
ax3.set_title("Iteration map")
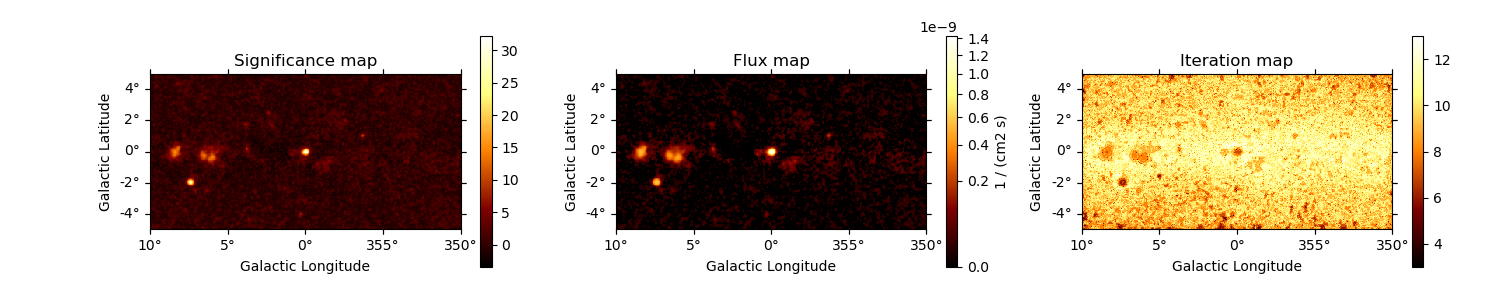
Text(0.5, 1.0, 'Iteration map')
Source candidates#
Let’s run a peak finder on the sqrt_ts image to get a list of
point-sources candidates (positions and peak sqrt_ts values). The
find_peaks function performs a local maximum search in a sliding
window, the argument min_distance is the minimum pixel distance
between peaks (smallest possible value and default is 1 pixel).
sources = find_peaks(maps["sqrt_ts"], threshold=5, min_distance="0.25 deg")
nsou = len(sources)
display(sources)
# Plot sources on top of significance sky image
plt.figure(figsize=(9, 5))
ax = maps["sqrt_ts"].plot(add_cbar=True)
ax.scatter(
sources["ra"],
sources["dec"],
transform=plt.gca().get_transform("icrs"),
color="none",
edgecolor="w",
marker="o",
s=600,
lw=1.5,
)
plt.show()
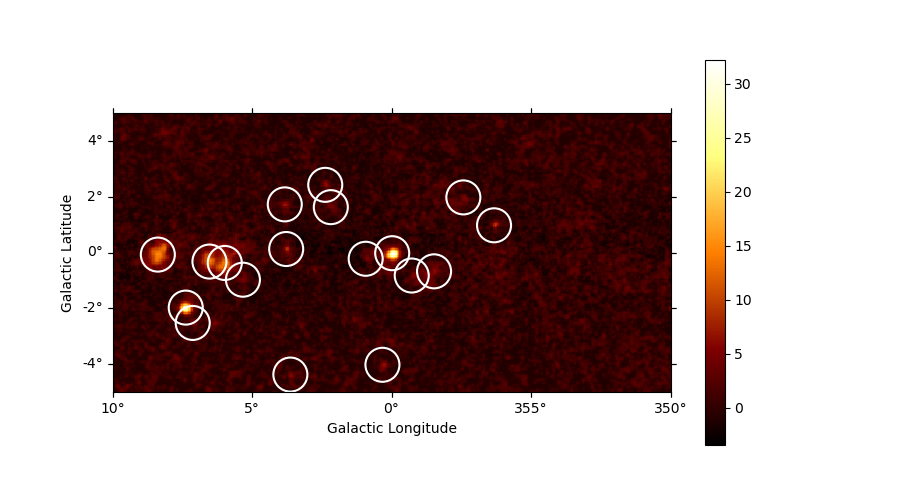
value x y ra dec
deg deg
------ --- --- --------- ---------
32.206 200 99 266.41449 -28.97054
27.836 52 60 272.43197 -23.54282
15.171 32 98 271.16056 -21.74479
14.143 69 93 270.40919 -23.47797
13.882 80 92 270.15899 -23.98049
9.7642 273 119 263.18257 -31.52587
8.7947 124 102 268.46711 -25.63326
7.3501 123 134 266.97596 -24.77174
6.8086 193 19 270.59696 -30.69138
6.2444 152 148 265.48068 -25.64323
5.8767 230 86 266.15140 -30.58926
5.6659 127 12 272.77351 -27.97934
5.6556 251 139 262.90685 -30.05853
5.4732 181 95 267.17020 -28.26173
5.4236 214 83 266.78188 -29.98429
5.1755 57 49 272.82739 -24.02653
5.0674 156 132 266.12148 -26.23306
5.0447 93 80 270.37773 -24.84233
Note that we used the instrument point-spread-function (PSF) as kernel, so the hypothesis we test is the presence of a point source. In order to test for extended sources we would have to use as kernel an extended template convolved by the PSF. Alternatively, we can compute the significance of an extended excess using the Li & Ma formalism, which is faster as no fitting is involve.
What next?#
In this notebook, we have seen how to work with images and compute TS and significance images from counts data, if a background estimate is already available.
Here’s some suggestions what to do next:
Look how background estimation is performed for IACTs with and without the high level interface in High level interface and Low level API notebooks, respectively
Learn about 2D model fitting in the 2D map fitting notebook
Find more about Fermi-LAT data analysis in the
:doc:/tutorials/data/fermi_lat` notebookUse source candidates to build a model and perform a 3D fitting (see 3D detailed analysis, Multi instrument joint 3D and 1D analysis notebooks for some hints)
Total running time of the script: ( 0 minutes 21.169 seconds)