Note
Go to the end to download the full example code or to run this example in your browser via Binder
Point source sensitivity#
Estimate the CTA sensitivity for a point-like IRF at a fixed zenith angle and fixed offset.
Introduction#
This notebook explains how to estimate the CTA sensitivity for a point-like IRF at a fixed zenith angle and fixed offset using the full containment IRFs distributed for the CTA 1DC. The significance is computed for a 1D analysis (On-OFF regions) and the LiMa formula.
We use here an approximate approach with an energy dependent integration radius to take into account the variation of the PSF. We will first determine the 1D IRFs including a containment correction.
We will be using the following Gammapy class:
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord
# %matplotlib inline
import matplotlib.pyplot as plt
Setup#
As usual, we’ll start with some setup …
from IPython.display import display
from gammapy.data import Observation, observatory_locations
from gammapy.datasets import SpectrumDataset, SpectrumDatasetOnOff
from gammapy.estimators import FluxPoints, SensitivityEstimator
from gammapy.irf import load_irf_dict_from_file
from gammapy.makers import SpectrumDatasetMaker
from gammapy.maps import MapAxis, RegionGeom
from gammapy.maps.axes import UNIT_STRING_FORMAT
Check setup#
from gammapy.utils.check import check_tutorials_setup
check_tutorials_setup()
System:
python_executable : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/bin/python
python_version : 3.9.16
machine : x86_64
system : Linux
Gammapy package:
version : 1.1
path : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/gammapy
Other packages:
numpy : 1.24.3
scipy : 1.10.1
astropy : 5.2.2
regions : 0.7
click : 8.1.3
yaml : 6.0
IPython : 8.14.0
jupyterlab : not installed
matplotlib : 3.7.1
pandas : not installed
healpy : 1.16.2
iminuit : 2.21.3
sherpa : 4.15.1
naima : 0.10.0
emcee : 3.1.4
corner : 2.2.2
ray : 2.5.0
Gammapy environment variables:
GAMMAPY_DATA : /home/runner/work/gammapy-docs/gammapy-docs/gammapy-datasets/1.1
Define analysis region and energy binning#
Here we assume a source at 0.5 degree from pointing position. We perform a simple energy independent extraction for now with a radius of 0.1 degree.
energy_axis = MapAxis.from_energy_bounds("0.03 TeV", "30 TeV", nbin=20)
energy_axis_true = MapAxis.from_energy_bounds(
"0.01 TeV", "100 TeV", nbin=100, name="energy_true"
)
geom = RegionGeom.create("icrs;circle(0, 0.5, 0.1)", axes=[energy_axis])
empty_dataset = SpectrumDataset.create(geom=geom, energy_axis_true=energy_axis_true)
Load IRFs and prepare dataset#
We extract the 1D IRFs from the full 3D IRFs provided by CTA.
irfs = load_irf_dict_from_file(
"$GAMMAPY_DATA/cta-1dc/caldb/data/cta/1dc/bcf/South_z20_50h/irf_file.fits"
)
location = observatory_locations["cta_south"]
pointing = SkyCoord("0 deg", "0 deg")
livetime = 5.0 * u.h
obs = Observation.create(
pointing=pointing, irfs=irfs, livetime=livetime, location=location
)
spectrum_maker = SpectrumDatasetMaker(selection=["exposure", "edisp", "background"])
dataset = spectrum_maker.run(empty_dataset, obs)
/home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/astropy/units/core.py:2097: UnitsWarning: '1/s/MeV/sr' did not parse as fits unit: Numeric factor not supported by FITS If this is meant to be a custom unit, define it with 'u.def_unit'. To have it recognized inside a file reader or other code, enable it with 'u.add_enabled_units'. For details, see https://docs.astropy.org/en/latest/units/combining_and_defining.html
warnings.warn(msg, UnitsWarning)
/home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/gammapy/data/observations.py:226: GammapyDeprecationWarning: Pointing will be required to be provided as FixedPointingInfo
warnings.warn(
Now we correct for the energy dependent region size:
containment = 0.68
# correct exposure
dataset.exposure *= containment
# correct background estimation
on_radii = obs.psf.containment_radius(
energy_true=energy_axis.center, offset=0.5 * u.deg, fraction=containment
)
factor = (1 - np.cos(on_radii)) / (1 - np.cos(geom.region.radius))
dataset.background *= factor.value.reshape((-1, 1, 1))
And finally define a SpectrumDatasetOnOff with an alpha of 0.2.
The off counts are created from the background model:
dataset_on_off = SpectrumDatasetOnOff.from_spectrum_dataset(
dataset=dataset, acceptance=1, acceptance_off=5
)
Compute sensitivity#
We impose a minimal number of expected signal counts of 5 per bin and a minimal significance of 3 per bin. We assume an alpha of 0.2 (ratio between ON and OFF area). We then run the sensitivity estimator.
sensitivity_estimator = SensitivityEstimator(
gamma_min=5, n_sigma=3, bkg_syst_fraction=0.10
)
sensitivity_table = sensitivity_estimator.run(dataset_on_off)
Results#
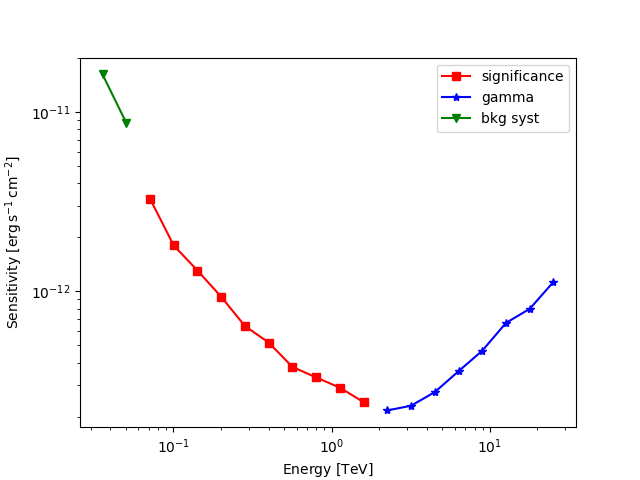
The results are given as an Astropy table. A column criterion allows to distinguish bins where the significance is limited by the signal statistical significance from bins where the sensitivity is limited by the number of signal counts. This is visible in the plot below.
# Show the results table
display(sensitivity_table)
# Save it to file (could use e.g. format of CSV or ECSV or FITS)
# sensitivity_table.write('sensitivity.ecsv', format='ascii.ecsv')
# Plot the sensitivity curve
t = sensitivity_table
is_s = t["criterion"] == "significance"
fig, ax = plt.subplots()
ax.plot(
t["e_ref"][is_s],
t["e2dnde"][is_s],
"s-",
color="red",
label="significance",
)
is_g = t["criterion"] == "gamma"
ax.plot(t["e_ref"][is_g], t["e2dnde"][is_g], "*-", color="blue", label="gamma")
is_bkg_syst = t["criterion"] == "bkg"
ax.plot(
t["e_ref"][is_bkg_syst],
t["e2dnde"][is_bkg_syst],
"v-",
color="green",
label="bkg syst",
)
ax.loglog()
ax.set_xlabel(f"Energy [{t['e_ref'].unit.to_string(UNIT_STRING_FORMAT)}]")
ax.set_ylabel(f"Sensitivity [{t['e2dnde'].unit.to_string(UNIT_STRING_FORMAT)}]")
ax.legend()
plt.show()
energy e_ref e_min e_max ... excess background criterion
TeV TeV TeV TeV ...
--------- --------- --------- --------- ... ------- ---------- ------------
0.0356551 0.0356551 0.03 0.0423761 ... 328.477 3284.77 bkg
0.0503641 0.0503641 0.0423761 0.0598579 ... 165.06 1650.6 bkg
0.0711412 0.0711412 0.0598579 0.0845515 ... 92.4984 756.756 significance
0.10049 0.10049 0.0845515 0.119432 ... 65.8637 376.563 significance
0.141945 0.141945 0.119432 0.168702 ... 46.0027 178.563 significance
0.200503 0.200503 0.168702 0.238298 ... 30.9746 77.2881 significance
0.283218 0.283218 0.238298 0.336606 ... 21.3831 34.5112 significance
0.400056 0.400056 0.336606 0.475468 ... 14.9743 15.4188 significance
0.565095 0.565095 0.475468 0.671616 ... 11.0135 7.41691 significance
0.798218 0.798218 0.671616 0.948683 ... 8.17611 3.4654 significance
1.12751 1.12751 0.948683 1.34005 ... 6.53986 1.86033 significance
1.59265 1.59265 1.34005 1.89287 ... 5.65742 1.19845 significance
2.24968 2.24968 1.89287 2.67375 ... 5 0.743113 gamma
3.17776 3.17776 2.67375 3.77678 ... 5 0.466104 gamma
4.48871 4.48871 3.77678 5.33484 ... 5 0.3196 gamma
6.34047 6.34047 5.33484 7.53566 ... 5 0.18669 gamma
8.95615 8.95615 7.53566 10.6444 ... 5 0.0845693 gamma
12.6509 12.6509 10.6444 15.0356 ... 5 0.0447552 gamma
17.8699 17.8699 15.0356 21.2384 ... 5 0.0266192 gamma
25.2419 25.2419 21.2384 30 ... 5 0.0126585 gamma
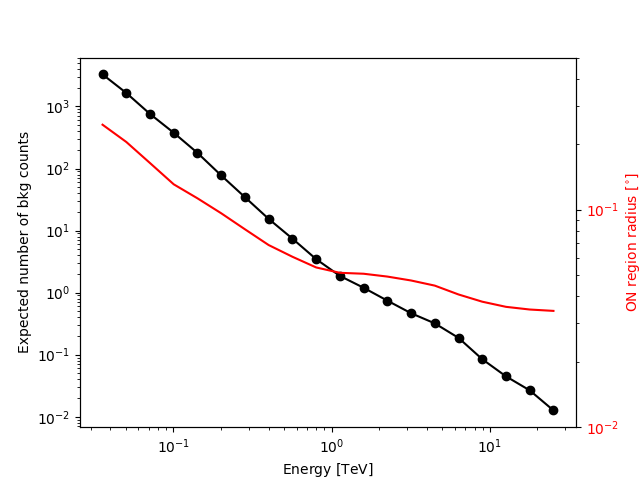
We add some control plots showing the expected number of background counts per bin and the ON region size cut (here the 68% containment radius of the PSF).
# Plot expected number of counts for signal and background
fig, ax1 = plt.subplots()
# ax1.plot( t["e_ref"], t["excess"],"o-", color="red", label="signal")
ax1.plot(t["e_ref"], t["background"], "o-", color="black", label="blackground")
ax1.loglog()
ax1.set_xlabel(f"Energy [{t['e_ref'].unit.to_string(UNIT_STRING_FORMAT)}]")
ax1.set_ylabel("Expected number of bkg counts")
ax2 = ax1.twinx()
ax2.set_ylabel(
f"ON region radius [{on_radii.unit.to_string(UNIT_STRING_FORMAT)}]", color="red"
)
ax2.semilogy(t["e_ref"], on_radii, color="red", label="PSF68")
ax2.tick_params(axis="y", labelcolor="red")
ax2.set_ylim(0.01, 0.5)
plt.show()
Obtaining an integral flux sensitivity#
It is often useful to obtain the integral sensitivity above a certain threshold. In this case, it is simplest to use a dataset with one energy bin while setting the high energy edge to a very large value. Here, we simply squash the previously created dataset into one with a single energy
dataset_on_off1 = dataset_on_off.to_image()
sensitivity_estimator = SensitivityEstimator(
gamma_min=5, n_sigma=3, bkg_syst_fraction=0.10
)
sensitivity_table = sensitivity_estimator.run(dataset_on_off1)
print(sensitivity_table)
# To get the integral flux, we convert to a `FluxPoints` object that does the conversion
# internally
flux_points = FluxPoints.from_table(
sensitivity_table, sed_type="e2dnde", reference_model=sensitivity_estimator.spectrum
)
print(
f"Integral sensitivity in {livetime:.2f} above {energy_axis.edges[0]:.2e} "
f"is {np.squeeze(flux_points.flux.quantity):.2e}"
)
energy e_ref e_min e_max e2dnde excess background criterion
TeV TeV TeV TeV erg / (cm2 s)
-------- -------- ----- ----- ------------- ------- ---------- ---------
0.948683 0.948683 0.03 30 1.44749e-12 639.029 6390.29 bkg
Integral sensitivity in 5.00 h above 3.00e-02 TeV is 3.01e-11 1 / (cm2 s)
Exercises#
Also compute the sensitivity for a 20 hour observation
Compare how the sensitivity differs between 5 and 20 hours by plotting the ratio as a function of energy.