This is a fixed-text formatted version of a Jupyter notebook
- Try online
- You can contribute with your own notebooks in this GitHub repository.
- Source files: spectrum_models.ipynb | spectrum_models.py
Spectral models in Gammapy¶
Introduction¶
This notebook explains how to use the functions and classes in gammapy.spectrum.models in order to work with spectral models.
The following clases will be used:
Setup¶
Same procedure as in every script …
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
[2]:
import numpy as np
import astropy.units as u
from gammapy.spectrum import models
from gammapy.utils.fitting import Parameter, Parameters
Create a model¶
To create a spectral model, instantiate an object of the spectral model class you’re interested in.
[3]:
pwl = models.PowerLaw()
print(pwl)
PowerLaw
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
index 2.000e+00 nan nan nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 nan nan False
reference 1.000e+00 nan TeV nan nan True
This will use default values for the model parameters, which is rarely what you want.
Usually you will want to specify the parameters on object creation. One way to do this is to pass astropy.utils.Quantity objects like this:
[4]:
pwl = models.PowerLaw(
index=2.3, amplitude=1e-12 * u.Unit("cm-2 s-1 TeV-1"), reference=1 * u.TeV
)
print(pwl)
PowerLaw
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
index 2.300e+00 nan nan nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 nan nan False
reference 1.000e+00 nan TeV nan nan True
As you see, some of the parameters have default min and values as well as a frozen flag. This is only relevant in the context of spectral fitting and thus covered in spectrum_analysis.ipynb. Also, the parameter errors are not set. This will be covered later in this tutorial.
Get and set model parameters¶
The model parameters are stored in the Parameters object on the spectal model. Each model parameter is a Parameter instance. It has a value and a unit attribute, as well as a quantity property for convenience.
[5]:
print(pwl.parameters)
Parameters
Parameter(name='index', value=2.3, factor=2.3, scale=1.0, unit='', min=nan, max=nan, frozen=False)
Parameter(name='amplitude', value=1e-12, factor=1e-12, scale=1.0, unit='cm-2 s-1 TeV-1', min=nan, max=nan, frozen=False)
Parameter(name='reference', value=1.0, factor=1.0, scale=1.0, unit='TeV', min=nan, max=nan, frozen=True)
covariance:
None
[6]:
print(pwl.parameters["index"])
pwl.parameters["index"].value = 2.6
print(pwl.parameters["index"])
Parameter(name='index', value=2.3, factor=2.3, scale=1.0, unit='', min=nan, max=nan, frozen=False)
Parameter(name='index', value=2.6, factor=2.6, scale=1.0, unit='', min=nan, max=nan, frozen=False)
[7]:
print(pwl.parameters["amplitude"])
pwl.parameters["amplitude"].quantity = 2e-12 * u.Unit("m-2 TeV-1 s-1")
print(pwl.parameters["amplitude"])
Parameter(name='amplitude', value=1e-12, factor=1e-12, scale=1.0, unit='cm-2 s-1 TeV-1', min=nan, max=nan, frozen=False)
Parameter(name='amplitude', value=2e-12, factor=2e-12, scale=1.0, unit='m-2 s-1 TeV-1', min=nan, max=nan, frozen=False)
List available models¶
All spectral models in gammapy are subclasses of SpectralModel. The list of available models is shown below.
[8]:
models.SpectralModel.__subclasses__()
[8]:
[gammapy.spectrum.models.ConstantModel,
gammapy.spectrum.models.CompoundSpectralModel,
gammapy.spectrum.models.PowerLaw,
gammapy.spectrum.models.PowerLaw2,
gammapy.spectrum.models.ExponentialCutoffPowerLaw,
gammapy.spectrum.models.ExponentialCutoffPowerLaw3FGL,
gammapy.spectrum.models.PLSuperExpCutoff3FGL,
gammapy.spectrum.models.LogParabola,
gammapy.spectrum.models.TableModel,
gammapy.spectrum.models.ScaleModel,
gammapy.spectrum.models.AbsorbedSpectralModel,
gammapy.spectrum.crab.MeyerCrabModel]
Plotting¶
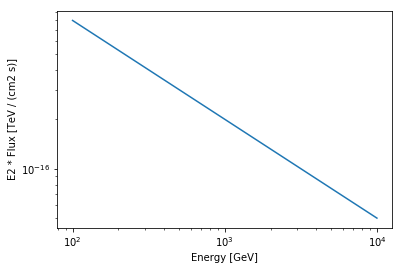
In order to plot a model you can use the plot function. It expects an energy range as argument. You can also chose flux and energy units as well as an energy power for the plot
[9]:
energy_range = [0.1, 10] * u.TeV
pwl.plot(energy_range, energy_power=2, energy_unit="GeV")
[9]:
<matplotlib.axes._subplots.AxesSubplot at 0x1a16a6c6d8>
Parameter errors¶
Parameters are stored internally as covariance matrix. There are, however, convenience methods to set individual parameter errors.
[10]:
pwl.parameters.set_parameter_errors(
{"index": 0.2, "amplitude": 0.1 * pwl.parameters["amplitude"].quantity}
)
print(pwl)
PowerLaw
Parameters:
name value error unit min max frozen
--------- --------- --------- ------------- --- --- ------
index 2.600e+00 2.000e-01 nan nan False
amplitude 2.000e-12 2.000e-13 m-2 s-1 TeV-1 nan nan False
reference 1.000e+00 0.000e+00 TeV nan nan True
Covariance:
name index amplitude reference
--------- --------- --------- ---------
index 4.000e-02 0.000e+00 0.000e+00
amplitude 0.000e+00 4.000e-26 0.000e+00
reference 0.000e+00 0.000e+00 0.000e+00
You can access the parameter errors like this
[11]:
pwl.parameters.covariance
[11]:
array([[4.e-02, 0.e+00, 0.e+00],
[0.e+00, 4.e-26, 0.e+00],
[0.e+00, 0.e+00, 0.e+00]])
[12]:
pwl.parameters.error("index")
[12]:
0.2
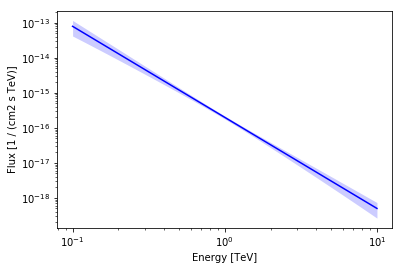
You can plot the butterfly using the plot_error method.
[13]:
ax = pwl.plot_error(energy_range, color="blue", alpha=0.2)
pwl.plot(energy_range, ax=ax, color="blue");
Integral fluxes¶
You’ve probably asked yourself already, if it’s possible to integrated models. Yes, it is. Where analytical solutions are available, these are used by default. Otherwise, a numerical integration is performed.
[14]:
pwl.integral(emin=1 * u.TeV, emax=10 * u.TeV)
[14]:
User-defined model¶
Now we’ll see how you can define a custom model. To do that you need to subclass SpectralModel. All SpectralModel subclasses need to have an __init__ function, which sets up the Parameters of the model and a static function called evaluate where the mathematical expression for the model is defined.
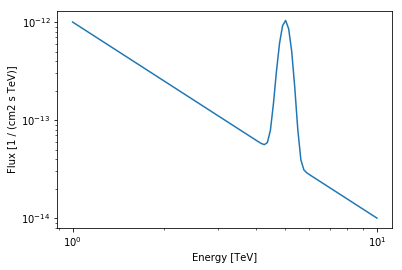
As an example we will use a PowerLaw plus a Gaussian (with fixed width).
[15]:
class UserModel(models.SpectralModel):
def __init__(self, index, amplitude, reference, mean, width):
self.parameters = Parameters(
[
Parameter("index", index, min=0),
Parameter("amplitude", amplitude, min=0),
Parameter("reference", reference, frozen=True),
Parameter("mean", mean, min=0),
Parameter("width", width, min=0, frozen=True),
]
)
@staticmethod
def evaluate(energy, index, amplitude, reference, mean, width):
pwl = models.PowerLaw.evaluate(
energy=energy,
index=index,
amplitude=amplitude,
reference=reference,
)
gauss = amplitude * np.exp(-(energy - mean) ** 2 / (2 * width ** 2))
return pwl + gauss
[16]:
model = UserModel(
index=2,
amplitude=1e-12 * u.Unit("cm-2 s-1 TeV-1"),
reference=1 * u.TeV,
mean=5 * u.TeV,
width=0.2 * u.TeV,
)
print(model)
UserModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --------- --- ------
index 2.000e+00 nan 0.000e+00 nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 0.000e+00 nan False
reference 1.000e+00 nan TeV nan nan True
mean 5.000e+00 nan TeV 0.000e+00 nan False
width 2.000e-01 nan TeV 0.000e+00 nan True
[17]:
energy_range = [1, 10] * u.TeV
model.plot(energy_range=energy_range);
What’s next?¶
In this tutorial we learnd how to work with spectral models.
Go to gammapy.spectrum to learn more.