This is a fixed-text formatted version of a Jupyter notebook
You may download all the notebooks as a tar file.
Source files: analysis_3d.ipynb | analysis_3d.py
3D detailed analysis#
This tutorial does a 3D map based analsis on the galactic center, using simulated observations from the CTA-1DC. We will use the high level interface for the data reduction, and then do a detailed modelling. This will be done in two different ways:
stacking all the maps together and fitting the stacked maps
handling all the observations separately and doing a joint fitting on all the maps
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import astropy.units as u
from pathlib import Path
from regions import CircleSkyRegion
from scipy.stats import norm
from gammapy.analysis import Analysis, AnalysisConfig
from gammapy.modeling.models import (
SkyModel,
ExpCutoffPowerLawSpectralModel,
PointSpatialModel,
FoVBackgroundModel,
Models,
)
from gammapy.modeling import Fit
from gammapy.maps import Map
from gammapy.estimators import ExcessMapEstimator
from gammapy.datasets import MapDataset
Analysis configuration#
In this section we select observations and define the analysis geometries, irrespective of joint/stacked analysis. For configuration of the analysis, we will programmatically build a config file from scratch.
[2]:
config = AnalysisConfig()
# The config file is now empty, with only a few defaults specified.
print(config)
AnalysisConfig
general:
log: {level: info, filename: null, filemode: null, format: null, datefmt: null}
outdir: .
n_jobs: 1
datasets_file: null
models_file: null
observations:
datastore: $GAMMAPY_DATA/hess-dl3-dr1
obs_ids: []
obs_file: null
obs_cone: {frame: null, lon: null, lat: null, radius: null}
obs_time: {start: null, stop: null}
required_irf: [aeff, edisp, psf, bkg]
datasets:
type: 1d
stack: true
geom:
wcs:
skydir: {frame: null, lon: null, lat: null}
binsize: 0.02 deg
width: {width: 5.0 deg, height: 5.0 deg}
binsize_irf: 0.2 deg
selection: {offset_max: 2.5 deg}
axes:
energy: {min: 1.0 TeV, max: 10.0 TeV, nbins: 5}
energy_true: {min: 0.5 TeV, max: 20.0 TeV, nbins: 16}
map_selection: [counts, exposure, background, psf, edisp]
background:
method: null
exclusion: null
parameters: {}
safe_mask:
methods: [aeff-default]
parameters: {}
on_region: {frame: null, lon: null, lat: null, radius: null}
containment_correction: true
fit:
fit_range: {min: null, max: null}
flux_points:
energy: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
excess_map:
correlation_radius: 0.1 deg
parameters: {}
energy_edges: {min: null, max: null, nbins: null}
light_curve:
time_intervals: {start: null, stop: null}
energy_edges: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
[3]:
# Selecting the observations
config.observations.datastore = "$GAMMAPY_DATA/cta-1dc/index/gps/"
config.observations.obs_ids = [110380, 111140, 111159]
[4]:
# Defining a reference geometry for the reduced datasets
config.datasets.type = "3d" # Analysis type is 3D
config.datasets.geom.wcs.skydir = {
"lon": "0 deg",
"lat": "0 deg",
"frame": "galactic",
} # The WCS geometry - centered on the galactic center
config.datasets.geom.wcs.width = {"width": "10 deg", "height": "8 deg"}
config.datasets.geom.wcs.binsize = "0.02 deg"
# Cutout size (for the run-wise event selection)
config.datasets.geom.selection.offset_max = 3.5 * u.deg
config.datasets.safe_mask.methods = ["aeff-default", "offset-max"]
# We now fix the energy axis for the counts map - (the reconstructed energy binning)
config.datasets.geom.axes.energy.min = "0.1 TeV"
config.datasets.geom.axes.energy.max = "10 TeV"
config.datasets.geom.axes.energy.nbins = 10
# We now fix the energy axis for the IRF maps (exposure, etc) - (the true energy binning)
config.datasets.geom.axes.energy_true.min = "0.08 TeV"
config.datasets.geom.axes.energy_true.max = "12 TeV"
config.datasets.geom.axes.energy_true.nbins = 14
[5]:
print(config)
AnalysisConfig
general:
log: {level: info, filename: null, filemode: null, format: null, datefmt: null}
outdir: .
n_jobs: 1
datasets_file: null
models_file: null
observations:
datastore: $GAMMAPY_DATA/cta-1dc/index/gps
obs_ids: [110380, 111140, 111159]
obs_file: null
obs_cone: {frame: null, lon: null, lat: null, radius: null}
obs_time: {start: null, stop: null}
required_irf: [aeff, edisp, psf, bkg]
datasets:
type: 3d
stack: true
geom:
wcs:
skydir: {frame: galactic, lon: 0.0 deg, lat: 0.0 deg}
binsize: 0.02 deg
width: {width: 10.0 deg, height: 8.0 deg}
binsize_irf: 0.2 deg
selection: {offset_max: 3.5 deg}
axes:
energy: {min: 0.1 TeV, max: 10.0 TeV, nbins: 10}
energy_true: {min: 0.08 TeV, max: 12.0 TeV, nbins: 14}
map_selection: [counts, exposure, background, psf, edisp]
background:
method: null
exclusion: null
parameters: {}
safe_mask:
methods: [aeff-default, offset-max]
parameters: {}
on_region: {frame: null, lon: null, lat: null, radius: null}
containment_correction: true
fit:
fit_range: {min: null, max: null}
flux_points:
energy: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
excess_map:
correlation_radius: 0.1 deg
parameters: {}
energy_edges: {min: null, max: null, nbins: null}
light_curve:
time_intervals: {start: null, stop: null}
energy_edges: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
Configuration for stacked and joint analysis#
This is done just by specfiying the flag on config.datasets.stack. Since the internal machinery will work differently for the two cases, we will write it as two config files and save it to disc in YAML format for future reference.
[6]:
config_stack = config.copy(deep=True)
config_stack.datasets.stack = True
config_joint = config.copy(deep=True)
config_joint.datasets.stack = False
[7]:
# To prevent unnecessary cluttering, we write it in a separate folder.
path = Path("analysis_3d")
path.mkdir(exist_ok=True)
config_joint.write(path=path / "config_joint.yaml", overwrite=True)
config_stack.write(path=path / "config_stack.yaml", overwrite=True)
Stacked analysis#
Data reduction#
We first show the steps for the stacked analysis and then repeat the same for the joint analysis later
[8]:
# Reading yaml file:
config_stacked = AnalysisConfig.read(path=path / "config_stack.yaml")
[9]:
analysis_stacked = Analysis(config_stacked)
Setting logging config: {'level': 'INFO', 'filename': None, 'filemode': None, 'format': None, 'datefmt': None}
[10]:
%%time
# select observations:
analysis_stacked.get_observations()
# run data reduction
analysis_stacked.get_datasets()
Fetching observations.
Observations selected: 3 out of 3.
Number of selected observations: 3
Creating reference dataset and makers.
Creating the background Maker.
No background maker set. Check configuration.
Start the data reduction loop.
Computing dataset for observation 110380
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 110380
No default lower safe energy threshold defined for obs 110380
Computing dataset for observation 111140
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 111140
No default lower safe energy threshold defined for obs 111140
Computing dataset for observation 111159
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 111159
No default lower safe energy threshold defined for obs 111159
CPU times: user 18.5 s, sys: 5.88 s, total: 24.4 s
Wall time: 27.8 s
We have one final dataset, which you can print and explore
[11]:
dataset_stacked = analysis_stacked.datasets["stacked"]
print(dataset_stacked)
MapDataset
----------
Name : stacked
Total counts : 121241
Total background counts : 108043.52
Total excess counts : 13197.48
Predicted counts : 108043.52
Predicted background counts : 108043.52
Predicted excess counts : nan
Exposure min : 6.28e+07 m2 s
Exposure max : 1.90e+10 m2 s
Number of total bins : 2000000
Number of fit bins : 1411180
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
[12]:
# To plot a smooth counts map
dataset_stacked.counts.smooth(0.02 * u.deg).plot_interactive(add_cbar=True)
[13]:
# And the background map
dataset_stacked.background.plot_interactive(add_cbar=True)
[14]:
# We can quickly check the PSF
dataset_stacked.psf.peek()
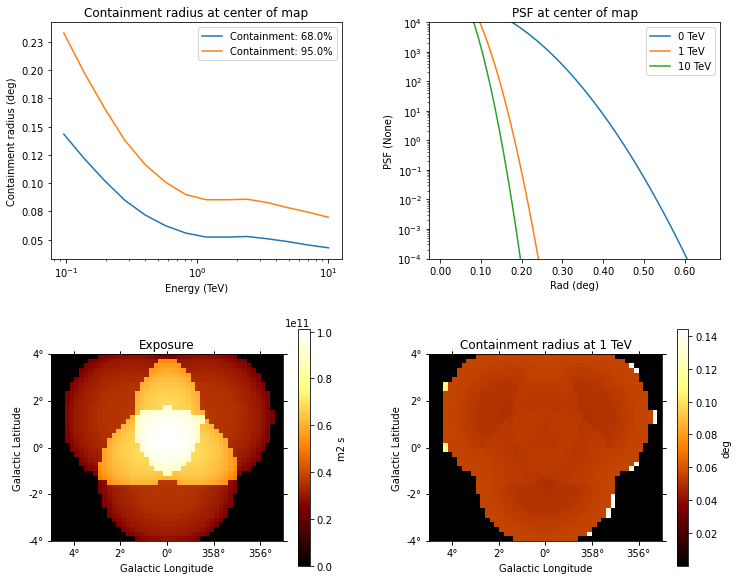
[15]:
# And the energy dispersion in the center of the map
dataset_stacked.edisp.peek()
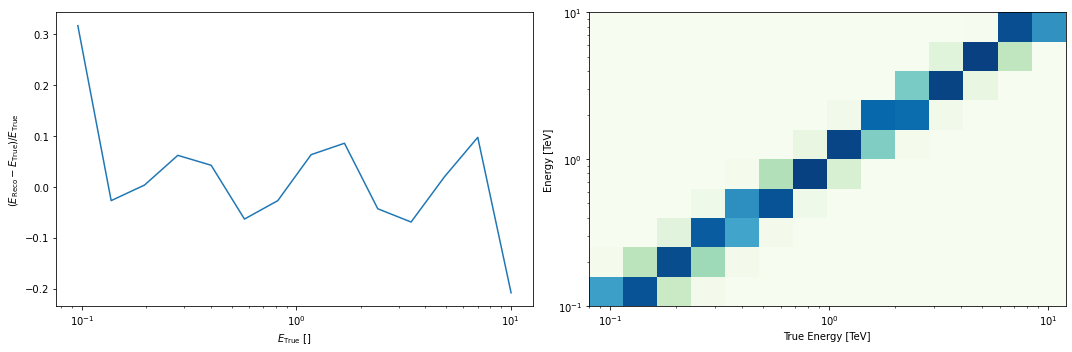
[16]:
# You can also get an excess image with a few lines of code:
excess = dataset_stacked.excess.sum_over_axes()
excess.smooth("0.06 deg").plot(stretch="sqrt", add_cbar=True);

Modeling and fitting#
Now comes the interesting part of the analysis - choosing appropriate models for our source and fitting them.
We choose a point source model with an exponential cutoff power-law spectrum.
To perform the fit on a restricted energy range, we can create a specific mask. On the dataset, the mask_fit is a Map sharing the same geometry as the MapDataset and containing boolean data.
To create a mask to limit the fit within a restricted energy range, one can rely on the gammapy.maps.Geom.energy_mask() method.
For more details on masks and the techniques to create them in gammapy, please refer to the dedicated tutorial
[17]:
dataset_stacked.mask_fit = dataset_stacked.counts.geom.energy_mask(
energy_min=0.3 * u.TeV, energy_max=None
)
[18]:
spatial_model = PointSpatialModel(
lon_0="-0.05 deg", lat_0="-0.05 deg", frame="galactic"
)
spectral_model = ExpCutoffPowerLawSpectralModel(
index=2.3,
amplitude=2.8e-12 * u.Unit("cm-2 s-1 TeV-1"),
reference=1.0 * u.TeV,
lambda_=0.02 / u.TeV,
)
model = SkyModel(
spatial_model=spatial_model,
spectral_model=spectral_model,
name="gc-source",
)
bkg_model = FoVBackgroundModel(dataset_name="stacked")
bkg_model.spectral_model.norm.value = 1.3
models_stacked = Models([model, bkg_model])
dataset_stacked.models = models_stacked
[19]:
%%time
fit = Fit(optimize_opts={"print_level": 1})
result = fit.run(datasets=[dataset_stacked])
CPU times: user 15.2 s, sys: 3.61 s, total: 18.8 s
Wall time: 19.9 s
Fit quality assessment and model residuals for a MapDataset#
We can access the results dictionary to see if the fit converged:
[20]:
print(result)
OptimizeResult
backend : minuit
method : migrad
success : True
message : Optimization terminated successfully.
nfev : 184
total stat : 180458.06
CovarianceResult
backend : minuit
method : hesse
success : True
message : Hesse terminated successfully.
Check best-fit parameters and error estimates:
[21]:
models_stacked.to_parameters_table()
[21]:
| model | type | name | value | unit | error | min | max | frozen | is_norm | link |
|---|---|---|---|---|---|---|---|---|---|---|
| str11 | str8 | str9 | float64 | str14 | float64 | float64 | float64 | bool | bool | str1 |
| gc-source | spectral | index | 2.4143e+00 | 1.523e-01 | nan | nan | False | False | ||
| gc-source | spectral | amplitude | 2.6631e-12 | cm-2 s-1 TeV-1 | 3.104e-13 | nan | nan | False | True | |
| gc-source | spectral | reference | 1.0000e+00 | TeV | 0.000e+00 | nan | nan | True | False | |
| gc-source | spectral | lambda_ | -1.3373e-02 | TeV-1 | 6.838e-02 | nan | nan | False | False | |
| gc-source | spectral | alpha | 1.0000e+00 | 0.000e+00 | nan | nan | True | False | ||
| gc-source | spatial | lon_0 | -4.8062e-02 | deg | 2.284e-03 | nan | nan | False | False | |
| gc-source | spatial | lat_0 | -5.2606e-02 | deg | 2.261e-03 | -9.000e+01 | 9.000e+01 | False | False | |
| stacked-bkg | spectral | norm | 1.3481e+00 | 9.315e-03 | nan | nan | False | True | ||
| stacked-bkg | spectral | tilt | 0.0000e+00 | 0.000e+00 | nan | nan | True | False | ||
| stacked-bkg | spectral | reference | 1.0000e+00 | TeV | 0.000e+00 | nan | nan | True | False |
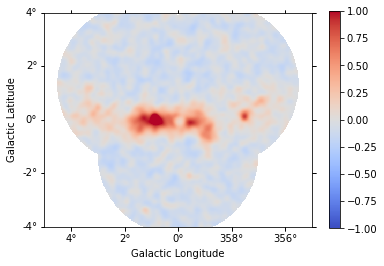
A quick way to inspect the model residuals is using the function ~MapDataset.plot_residuals_spatial(). This function computes and plots a residual image (by default, the smoothing radius is 0.1 deg and method=diff, which corresponds to a simple data - model plot):
[22]:
dataset_stacked.plot_residuals_spatial(
method="diff/sqrt(model)", vmin=-1, vmax=1
);
The more general function ~MapDataset.plot_residuals() can also extract and display spectral residuals in a region:
[23]:
region = CircleSkyRegion(spatial_model.position, radius=0.15 * u.deg)
dataset_stacked.plot_residuals(
kwargs_spatial=dict(method="diff/sqrt(model)", vmin=-1, vmax=1),
kwargs_spectral=dict(region=region),
);
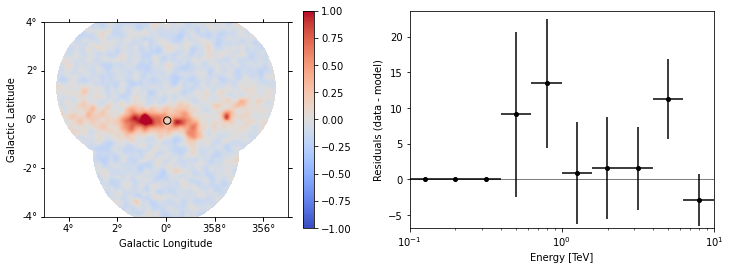
This way of accessing residuals is quick and handy, but comes with limitations. For example: - In case a fitting energy range was defined using a MapDataset.mask_fit, it won’t be taken into account. Residuals will be summed up over the whole reconstructed energy range - In order to make a proper statistic treatment, instead of simple residuals a proper residuals significance map should be computed
A more accurate way to inspect spatial residuals is the following:
[24]:
estimator = ExcessMapEstimator(
correlation_radius="0.1 deg",
selection_optional=[],
energy_edges=[0.1, 1, 10] * u.TeV,
)
result = estimator.run(dataset_stacked)
result["sqrt_ts"].plot_grid(
figsize=(12, 4), cmap="coolwarm", add_cbar=True, vmin=-5, vmax=5, ncols=2
);
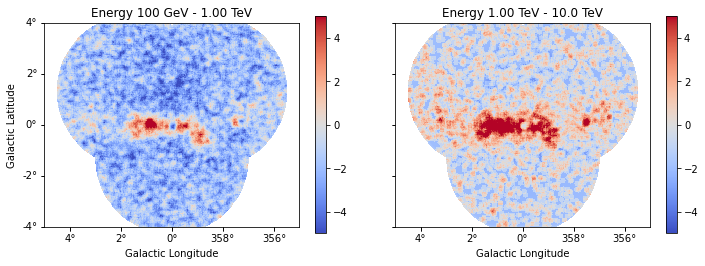
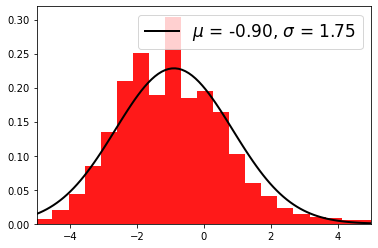
Distribution of residuals significance in the full map geometry:
[25]:
# TODO: clean this up
significance_data = result["sqrt_ts"].data
# #Remove bins that are inside an exclusion region, that would create an artificial peak at significance=0.
# #For now these lines are useless, because to_image() drops the mask fit
# mask_data = dataset_image.mask_fit.sum_over_axes().data
# excluded = mask_data == 0
# significance_data = significance_data[~excluded]
selection = np.isfinite(significance_data) & ~(significance_data == 0)
significance_data = significance_data[selection]
plt.hist(significance_data, density=True, alpha=0.9, color="red", bins=40)
mu, std = norm.fit(significance_data)
x = np.linspace(-5, 5, 100)
p = norm.pdf(x, mu, std)
plt.plot(
x,
p,
lw=2,
color="black",
label=r"$\mu$ = {:.2f}, $\sigma$ = {:.2f}".format(mu, std),
)
plt.legend(fontsize=17)
plt.xlim(-5, 5)
[25]:
(-5.0, 5.0)
Joint analysis#
In this section, we perform a joint analysis of the same data. Of course, joint fitting is considerably heavier than stacked one, and should always be handled with care. For brevity, we only show the analysis for a point source fitting without re-adding a diffuse component again.
Data reduction#
[26]:
%%time
# Read the yaml file from disk
config_joint = AnalysisConfig.read(path=path / "config_joint.yaml")
analysis_joint = Analysis(config_joint)
# select observations:
analysis_joint.get_observations()
# run data reduction
analysis_joint.get_datasets()
Setting logging config: {'level': 'INFO', 'filename': None, 'filemode': None, 'format': None, 'datefmt': None}
Fetching observations.
Observations selected: 3 out of 3.
Number of selected observations: 3
Creating reference dataset and makers.
Creating the background Maker.
No background maker set. Check configuration.
Start the data reduction loop.
Computing dataset for observation 110380
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 110380
No default lower safe energy threshold defined for obs 110380
Computing dataset for observation 111140
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 111140
No default lower safe energy threshold defined for obs 111140
Computing dataset for observation 111159
Running MapDatasetMaker
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Running SafeMaskMaker
No default upper safe energy threshold defined for obs 111159
No default lower safe energy threshold defined for obs 111159
CPU times: user 17.1 s, sys: 5.48 s, total: 22.5 s
Wall time: 23.8 s
[27]:
# You can see there are 3 datasets now
print(analysis_joint.datasets)
Datasets
--------
Dataset 0:
Type : MapDataset
Name : lGuyz0va
Instrument : CTA
Models :
Dataset 1:
Type : MapDataset
Name : cQIq0S4b
Instrument : CTA
Models :
Dataset 2:
Type : MapDataset
Name : K0C_ygHS
Instrument : CTA
Models :
[28]:
# You can access each one by name or by index, eg:
print(analysis_joint.datasets[0])
MapDataset
----------
Name : lGuyz0va
Total counts : 40481
Total background counts : 36014.51
Total excess counts : 4466.49
Predicted counts : 36014.51
Predicted background counts : 36014.51
Predicted excess counts : nan
Exposure min : 6.28e+07 m2 s
Exposure max : 6.68e+09 m2 s
Number of total bins : 1085000
Number of fit bins : 693940
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
After the data reduction stage, it is nice to get a quick summary info on the datasets. Here, we look at the statistics in the center of Map, by passing an appropriate region. To get info on the entire spatial map, omit the region argument.
[29]:
analysis_joint.datasets.info_table()
[29]:
| name | counts | excess | sqrt_ts | background | npred | npred_background | npred_signal | exposure_min | exposure_max | livetime | ontime | counts_rate | background_rate | excess_rate | n_bins | n_fit_bins | stat_type | stat_sum |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m2 s | m2 s | s | s | 1 / s | 1 / s | 1 / s | ||||||||||||
| str8 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | int64 | int64 | str4 | float64 |
| lGuyz0va | 40481 | 4466.484375 | 23.07275543163338 | 36014.515625 | 36014.50696021132 | 36014.515625 | nan | 62838924.0 | 6676807680.0 | 1764.000034326 | 1800.0 | 22.948412251855327 | 20.416391680378084 | 2.5320205714772457 | 1085000 | 693940 | cash | nan |
| cQIq0S4b | 40525 | 4510.505523195912 | 23.295778288208588 | 36014.49447680409 | 36014.49447680409 | 36014.49447680409 | nan | 62838194.40984807 | 6676808531.804668 | 1764.000034326 | 1800.0 | 22.97335556202755 | 20.416379691605123 | 2.5569758704224252 | 1085000 | 693940 | cash | nan |
| K0C_ygHS | 40235 | 4220.480554966161 | 21.82496424955487 | 36014.51944503384 | 36014.51944503384 | 36014.51944503384 | nan | 62838404.53574324 | 6676813626.917053 | 1764.000034326 | 1800.0 | 22.8089564722561 | 20.416393845930102 | 2.392562626325996 | 1085000 | 693940 | cash | nan |
[30]:
models_joint = Models()
model_joint = model.copy(name="source-joint")
models_joint.append(model_joint)
for dataset in analysis_joint.datasets:
bkg_model = FoVBackgroundModel(dataset_name=dataset.name)
models_joint.append(bkg_model)
print(models_joint)
Models
Component 0: SkyModel
Name : source-joint
Datasets names : None
Spectral model type : ExpCutoffPowerLawSpectralModel
Spatial model type : PointSpatialModel
Temporal model type :
Parameters:
index : 2.414 +/- 0.15
amplitude : 2.66e-12 +/- 3.1e-13 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
lambda_ : -0.013 +/- 0.07 1 / TeV
alpha (frozen): 1.000
lon_0 : -0.048 +/- 0.00 deg
lat_0 : -0.053 +/- 0.00 deg
Component 1: FoVBackgroundModel
Name : lGuyz0va-bkg
Datasets names : ['lGuyz0va']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
Component 2: FoVBackgroundModel
Name : cQIq0S4b-bkg
Datasets names : ['cQIq0S4b']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
Component 3: FoVBackgroundModel
Name : K0C_ygHS-bkg
Datasets names : ['K0C_ygHS']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
[31]:
# and set the new model
analysis_joint.datasets.models = models_joint
[32]:
%%time
fit_joint = Fit()
result_joint = fit_joint.run(datasets=analysis_joint.datasets)
CPU times: user 30 s, sys: 7.28 s, total: 37.3 s
Wall time: 41.8 s
Fit quality assessment and model residuals for a joint Datasets#
We can access the results dictionary to see if the fit converged:
[33]:
print(result_joint)
OptimizeResult
backend : minuit
method : migrad
success : True
message : Optimization terminated successfully.
nfev : 227
total stat : 748259.16
CovarianceResult
backend : minuit
method : hesse
success : True
message : Hesse terminated successfully.
Check best-fit parameters and error estimates:
[34]:
print(models_joint)
Models
Component 0: SkyModel
Name : source-joint
Datasets names : None
Spectral model type : ExpCutoffPowerLawSpectralModel
Spatial model type : PointSpatialModel
Temporal model type :
Parameters:
index : 2.272 +/- 0.08
amplitude : 2.84e-12 +/- 3.1e-13 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
lambda_ : 0.039 +/- 0.05 1 / TeV
alpha (frozen): 1.000
lon_0 : -0.049 +/- 0.00 deg
lat_0 : -0.053 +/- 0.00 deg
Component 1: FoVBackgroundModel
Name : lGuyz0va-bkg
Datasets names : ['lGuyz0va']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.118 +/- 0.01
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
Component 2: FoVBackgroundModel
Name : cQIq0S4b-bkg
Datasets names : ['cQIq0S4b']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.119 +/- 0.01
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
Component 3: FoVBackgroundModel
Name : K0C_ygHS-bkg
Datasets names : ['K0C_ygHS']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.111 +/- 0.01
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
Since the joint dataset is made of multiple datasets, we can either: - Look at the residuals for each dataset separately. In this case, we can directly refer to the section Fit quality and model residuals for a MapDataset in this notebook - Look at a stacked residual map.
In the latter case, we need to properly stack the joint dataset before computing the residuals:
[35]:
# TODO: clean this up
# We need to stack on the full geometry, so we use to geom from the stacked counts map.
stacked = MapDataset.from_geoms(**dataset_stacked.geoms)
for dataset in analysis_joint.datasets:
# TODO: Apply mask_fit before stacking
stacked.stack(dataset)
stacked.models = [model_joint]
[36]:
stacked.plot_residuals_spatial(vmin=-1, vmax=1);
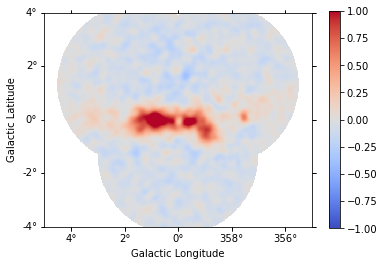
Then, we can access the stacked model residuals as previously shown in the section Fit quality and model residuals for a MapDataset in this notebook.
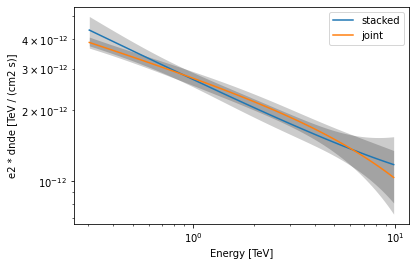
Finally, let us compare the spectral results from the stacked and joint fit:
[37]:
def plot_spectrum(model, result, label, color):
spec = model.spectral_model
energy_bounds = [0.3, 10] * u.TeV
spec.plot(
energy_bounds=energy_bounds, energy_power=2, label=label, color=color
)
spec.plot_error(energy_bounds=energy_bounds, energy_power=2, color=color)
[38]:
plot_spectrum(model, result, label="stacked", color="tab:blue")
plot_spectrum(model_joint, result_joint, label="joint", color="tab:orange")
plt.legend()
[38]:
<matplotlib.legend.Legend at 0x146311d30>
Summary#
Note that this notebook aims to show you the procedure of a 3D analysis using just a few observations. Results get much better for a more complete analysis considering the GPS dataset from the CTA First Data Challenge (DC-1) and also the CTA model for the Galactic diffuse emission, as shown in the next image:
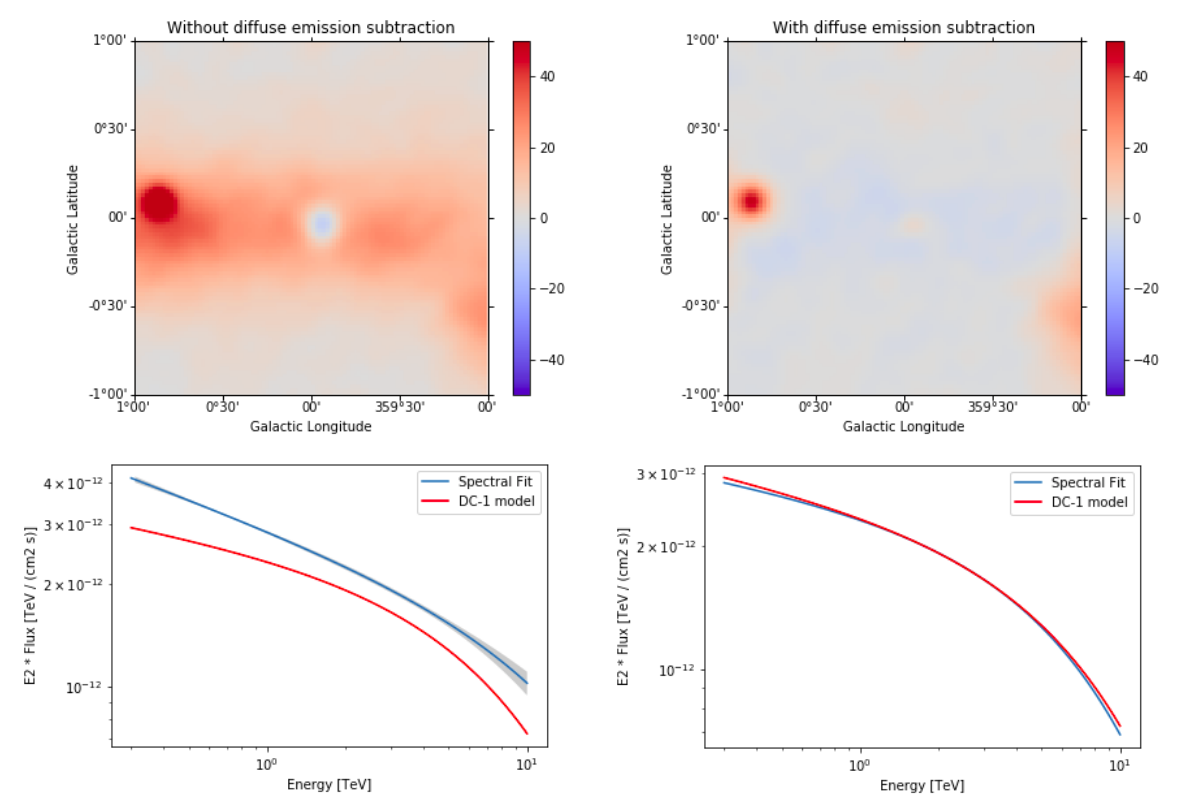
The complete tutorial notebook of this analysis is available to be downloaded in GAMMAPY-EXTRA repository at https://github.com/gammapy/gammapy-extra/blob/master/analyses/cta_1dc_gc_3d.ipynb).
Exercises#
Analyse the second source in the field of view: G0.9+0.1 and add it to the combined model.
Perform modeling in more details - Add diffuse component, get flux points.
