Note
Go to the end to download the full example code or to run this example in your browser via Binder
Observational clustering#
Clustering observations into specific groups.
Context#
Typically, observations from gamma-ray telescopes can span a number of different observation periods, therefore it is likely that the observation conditions and quality are not always the same. This tutorial aims to provide a way in which observations can be grouped such that similar observations are grouped together, and then the data reduction is performed.
Objective#
To cluster similar observations based on various quantities.
Proposed approach#
For completeness two different methods for grouping of observations are shown here.
A simple grouping based on zenith angle from an existing observations table.
Grouping the observations depending on the IRF quality, by means of hierarchical clustering.
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord
import matplotlib.pyplot as plt
from gammapy.data import DataStore
from gammapy.data.observations import Observations
from gammapy.data.utils import get_irfs_features
from gammapy.utils.cluster import hierarchical_clustering, standard_scaler
Obtain the observations#
First need to define the DataStore object for the HESS DL3 DR1
data. Next, utilise a cone search to select only the observations of interest.
In this case, we choose PKS 2155-304 as the object of interest.
The ObservationTable is then filtered using the select_observations tool.
data_store = DataStore.from_dir("$GAMMAPY_DATA/hess-dl3-dr1")
selection = dict(
type="sky_circle",
frame="icrs",
lon="329.71693826 deg",
lat="-30.2255 deg",
radius="2 deg",
)
obs_table = data_store.obs_table
selected_obs_table = obs_table.select_observations(selection)
More complex selection can be done by utilising the obs_table entries directly.
We can now retrieve the relevant observations by passing their obs_id to the
get_observations method.
obs_ids = selected_obs_table["OBS_ID"]
observations = data_store.get_observations(obs_ids)
Show various observation quantities#
Print here the range of zenith angles and muon efficiencies, to see if there is a sensible way to group the observations.
obs_zenith = selected_obs_table["ZEN_PNT"].to(u.deg)
obs_muoneff = selected_obs_table["MUONEFF"]
print(f"{np.min(obs_zenith):.2f} < zenith angle < {np.max(obs_zenith):.2f}")
print(f"{np.min(obs_muoneff):.2f} < muon efficiency < {np.max(obs_muoneff):.2f}")
7.23 deg < zenith angle < 50.42 deg
0.97 < muon efficiency < 1.03
Manual grouping of observations#
Here we can plot the zenith angle vs muon efficiency of the observations. We decide to group the observations according to their zenith angle. This is done manually as per a user defined cut, in this case we take the median value of the zenith angles to define each observation group.
This type of grouping can be utilised according to different parameters i.e. zenith angle, muon efficiency, offset angle. The quantity chosen can therefore be adjusted according to each specific science case.
median_zenith = np.median(obs_zenith)
labels = []
for obs in observations:
zenith = obs.get_pointing_altaz(time=obs.tmid).zen
labels.append("low_zenith" if zenith < median_zenith else "high_zenith")
grouped_observations = observations.group_by_label(labels)
print(grouped_observations)
{'group_high_zenith': <gammapy.data.observations.Observations object at 0x7f1564d3eb50>, 'group_low_zenith': <gammapy.data.observations.Observations object at 0x7f15683af9a0>}
The results for each group of observations is shown visually below.
fix, ax = plt.subplots(1, 1, figsize=(7, 5))
for obs in grouped_observations["group_low_zenith"]:
ax.plot(
obs.get_pointing_altaz(time=obs.tmid).zen,
obs.meta.optional["MUONEFF"],
"d",
color="red",
)
for obs in grouped_observations["group_high_zenith"]:
ax.plot(
obs.get_pointing_altaz(time=obs.tmid).zen,
obs.meta.optional["MUONEFF"],
"o",
color="blue",
)
ax.set_ylabel("Muon efficiency")
ax.set_xlabel("Zenith angle (deg)")
ax.axvline(median_zenith.value, ls="--", color="black")
plt.show()
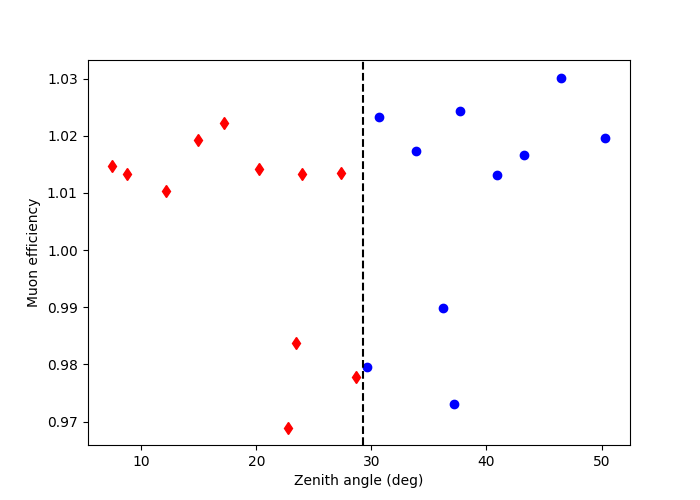
This shows the observations grouped by zenith angle. The diamonds are observations which have a zenith angle less than the median value, whilst the circles are observations above the median.
The grouped_observations provide a list of Observations
which can be utilised in the usual way to show the various properties
of the observations i.e. see the CTA with Gammapy tutorial.
Hierarchical clustering of observations#
This method shows how to cluster observations based on their IRF quantities,
in this case those that have a similar edisp and psf. The
get_irfs_features is utilised to achieve this. The
observations are then clustered based on these criteria using
hierarchical_clustering. The idea here is to minimise
the variance of both edisp and psf within a specific group to limit the error
on the quantity when they are stacked at the dataset level.
In this example, the irf features are computed for the edisp-res and
psf-radius at 1 TeV. This is stored as a Table, as shown below.
source_position = SkyCoord(329.71693826 * u.deg, -30.2255890 * u.deg, frame="icrs")
names = ["edisp-res", "psf-radius"]
features_irfs = get_irfs_features(
observations, energy_true="1 TeV", position=source_position, names=names
)
print(features_irfs)
edisp-res obs_id psf-radius
deg
------------------- ------ -------------------
0.36834038301420274 33787 0.14149953611195087
0.339835555384604 33788 0.11553325504064559
0.3237948931463171 33789 0.10262943822890519
0.30535345877453707 33790 0.09426693227142095
0.28755283551095173 33791 0.08894569035619496
0.27409496735322464 33792 0.08447355125099419
0.26665050077722524 33793 0.0811551760882139
0.26272868097919794 33794 0.07943648658692837
0.2639554729438709 33795 0.0799109224230051
0.26887783978974283 33796 0.08191603310406206
0.2777074437073429 33797 0.0855013383552432
0.29355238360800506 33798 0.0897868126630783
0.31186857659616535 33799 0.09623312838375568
0.33164865722698683 33800 0.10470702368766069
0.3503706026275275 33801 0.12276676166802643
0.3011061699260256 47802 0.09740295372903346
0.2861432787940619 47803 0.08880368117243051
0.27057337686547633 47804 0.08388624433428049
0.29882214027996945 47827 0.09610314778983592
0.28385358839966657 47828 0.08795162606984375
0.268663733018811 47829 0.08328557573258877
Compute standardized features by removing the mean and scaling to unit variance:
edisp-res obs_id psf-radius
-------------------- ------ ---------------------
2.3885947175689592 33787 3.053212009682775
1.4351637481047363 33788 1.363472509034498
0.8986348363207728 33789 0.5237647004325865
0.2818047723094509 33790 -0.020420144596410953
-0.3135914081482271 33791 -0.36669663417038234
-0.7637308880733709 33792 -0.6577182894355391
-1.012733796525585 33793 -0.873659477745188
-1.1439110308062257 33794 -0.985502122122975
-1.102877228833871 33795 -0.9546285068162436
-0.9382336444241555 33796 -0.8241471833009617
-0.6429005895278312 33797 -0.590835686434463
-0.11291820875721864 33798 -0.3119611261122878
0.49972277488662115 33799 0.10752883769757363
1.1613279491744304 33800 0.6589622747787678
1.7875405607868806 33801 1.8341884287660133
0.1397412321592923 47802 0.1836544903987521
-0.36073833513766157 47803 -0.3759377929871826
-0.8815212313941426 47804 -0.6959369197218669
0.06334488877417636 47827 0.09907043184188653
-0.4373240195300975 47828 -0.4313847458879893
-0.9453950989269149 47829 -0.7350250533013533
The hierarchical_clustering then clusters
this table into t=2 groups with a corresponding label for each group.
In this case, we choose to cluster the observations into two groups.
We can print this table to show the corresponding label which has been
added to the previous feature_irfs table.
The arguments for fcluster are passed as
a dictionary here.
features = hierarchical_clustering(scaled_features_irfs, fcluster_kwargs={"t": 2})
print(features)
edisp-res obs_id psf-radius labels
-------------------- ------ --------------------- ------
2.3885947175689592 33787 3.053212009682775 1
1.4351637481047363 33788 1.363472509034498 1
0.8986348363207728 33789 0.5237647004325865 1
0.2818047723094509 33790 -0.020420144596410953 2
-0.3135914081482271 33791 -0.36669663417038234 2
-0.7637308880733709 33792 -0.6577182894355391 2
-1.012733796525585 33793 -0.873659477745188 2
-1.1439110308062257 33794 -0.985502122122975 2
-1.102877228833871 33795 -0.9546285068162436 2
-0.9382336444241555 33796 -0.8241471833009617 2
-0.6429005895278312 33797 -0.590835686434463 2
-0.11291820875721864 33798 -0.3119611261122878 2
0.49972277488662115 33799 0.10752883769757363 2
1.1613279491744304 33800 0.6589622747787678 1
1.7875405607868806 33801 1.8341884287660133 1
0.1397412321592923 47802 0.1836544903987521 2
-0.36073833513766157 47803 -0.3759377929871826 2
-0.8815212313941426 47804 -0.6959369197218669 2
0.06334488877417636 47827 0.09907043184188653 2
-0.4373240195300975 47828 -0.4313847458879893 2
-0.9453950989269149 47829 -0.7350250533013533 2
Finally, observations.group_by_label creates a dictionary containing t
Observations objects by grouping the similar labels.
obs_clusters = observations.group_by_label(features["labels"])
print(obs_clusters)
mask_1 = features["labels"] == 1
mask_2 = features["labels"] == 2
fix, ax = plt.subplots(1, 1, figsize=(7, 5))
ax.set_ylabel("edisp-res")
ax.set_xlabel("psf-radius")
ax.plot(
features_irfs[mask_1]["edisp-res"],
features_irfs[mask_1]["psf-radius"],
"d",
color="green",
label="Group 1",
)
ax.plot(
features_irfs[mask_2]["edisp-res"],
features_irfs[mask_2]["psf-radius"],
"o",
color="magenta",
label="Group 2",
)
ax.legend()
plt.show()
{'group_1': <gammapy.data.observations.Observations object at 0x7f1562068280>, 'group_2': <gammapy.data.observations.Observations object at 0x7f1562068220>}
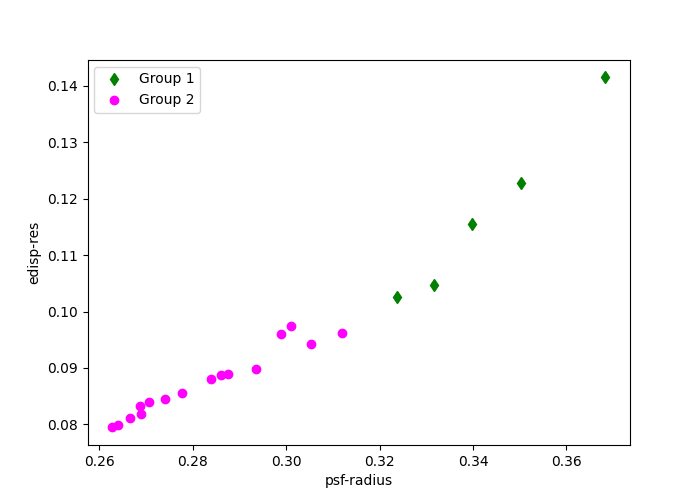
The groups here are divided by the quality of the IRFs values edisp-res
and psf-radius. The diamond and circular points indicate how the observations
are grouped.
In both examples we have a set of Observation objects which
can be reduced using the DatasetsMaker to create two (in this
specific case) separate datasets. These can then be jointly fitted using the
Multi instrument joint 3D and 1D analysis tutorial.