Note
Go to the end to download the full example code or to run this example in your browser via Binder
Datasets - Reduced data, IRFs, models#
Learn how to work with datasets
Introduction#
datasets are a crucial part of the gammapy API. Dataset
objects constitute DL4 data - binned counts, IRFs, models and the associated
likelihoods. Datasets from the end product of the data reduction stage,
see Makers - Data reduction tutorial and are passed on to the Fit
or estimator classes for modelling and fitting purposes.
To find the different types of Dataset objects that are supported see
Types of supported datasets:
Setup#
import astropy.units as u
from astropy.coordinates import SkyCoord
from regions import CircleSkyRegion
import matplotlib.pyplot as plt
from IPython.display import display
from gammapy.data import GTI
from gammapy.datasets import (
Datasets,
FluxPointsDataset,
MapDataset,
SpectrumDatasetOnOff,
)
from gammapy.estimators import FluxPoints
from gammapy.maps import MapAxis, WcsGeom
from gammapy.modeling.models import FoVBackgroundModel, PowerLawSpectralModel, SkyModel
Check setup#
from gammapy.utils.check import check_tutorials_setup
from gammapy.utils.scripts import make_path
# %matplotlib inline
check_tutorials_setup()
System:
python_executable : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/bin/python
python_version : 3.9.18
machine : x86_64
system : Linux
Gammapy package:
version : 1.2
path : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/gammapy
Other packages:
numpy : 1.26.4
scipy : 1.12.0
astropy : 5.2.2
regions : 0.8
click : 8.1.7
yaml : 6.0.1
IPython : 8.18.1
jupyterlab : not installed
matplotlib : 3.8.3
pandas : not installed
healpy : 1.16.6
iminuit : 2.25.2
sherpa : 4.16.0
naima : 0.10.0
emcee : 3.1.4
corner : 2.2.2
ray : 2.9.3
Gammapy environment variables:
GAMMAPY_DATA : /home/runner/work/gammapy-docs/gammapy-docs/gammapy-datasets/1.2
MapDataset#
The counts, exposure, background, masks, and IRF maps are bundled
together in a data structure named MapDataset. While the counts,
and background maps are binned in reconstructed energy and must have
the same geometry, the IRF maps can have a different spatial (coarsely
binned and larger) geometry and spectral range (binned in true
energies). It is usually recommended that the true energy bin should be
larger and more finely sampled and the reco energy bin.
Creating an empty dataset#
An empty MapDataset can be directly instantiated from any
WcsGeom object:
energy_axis = MapAxis.from_energy_bounds(1, 10, nbin=11, name="energy", unit="TeV")
geom = WcsGeom.create(
skydir=(83.63, 22.01),
axes=[energy_axis],
width=5 * u.deg,
binsz=0.05 * u.deg,
frame="icrs",
)
dataset_empty = MapDataset.create(geom=geom, name="my-dataset")
It is good practice to define a name for the dataset, such that you can identify it later by name. However if you define a name it must be unique. Now we can already print the dataset:
print(dataset_empty)
MapDataset
----------
Name : my-dataset
Total counts : 0
Total background counts : 0.00
Total excess counts : 0.00
Predicted counts : 0.00
Predicted background counts : 0.00
Predicted excess counts : nan
Exposure min : 0.00e+00 m2 s
Exposure max : 0.00e+00 m2 s
Number of total bins : 110000
Number of fit bins : 0
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
The printout shows the key summary information of the dataset, such as total counts, fit statistics, model information etc.
from_geom has additional keywords, that can be used to
define the binning of the IRF related maps:
# choose a different true energy binning for the exposure, psf and edisp
energy_axis_true = MapAxis.from_energy_bounds(
0.1, 100, nbin=11, name="energy_true", unit="TeV", per_decade=True
)
# choose a different rad axis binning for the psf
rad_axis = MapAxis.from_bounds(0, 5, nbin=50, unit="deg", name="rad")
gti = GTI.create(0 * u.s, 1000 * u.s)
dataset_empty = MapDataset.create(
geom=geom,
energy_axis_true=energy_axis_true,
rad_axis=rad_axis,
binsz_irf=0.1,
gti=gti,
name="dataset-empty",
)
To see the geometry of each map, we can use:
print(dataset_empty.geoms)
{'geom': <gammapy.maps.wcs.geom.WcsGeom object at 0x7f1567e81f40>, 'geom_exposure': <gammapy.maps.wcs.geom.WcsGeom object at 0x7f1566fe93a0>, 'geom_psf': <gammapy.maps.wcs.geom.WcsGeom object at 0x7f15697d72b0>, 'geom_edisp': <gammapy.maps.wcs.geom.WcsGeom object at 0x7f1564c99be0>}
Another way to create a MapDataset is to just read an existing one
from a FITS file:
dataset_cta = MapDataset.read(
"$GAMMAPY_DATA/cta-1dc-gc/cta-1dc-gc.fits.gz", name="dataset-cta"
)
print(dataset_cta)
MapDataset
----------
Name : dataset-cta
Total counts : 104317
Total background counts : 91507.70
Total excess counts : 12809.30
Predicted counts : 91507.69
Predicted background counts : 91507.70
Predicted excess counts : nan
Exposure min : 6.28e+07 m2 s
Exposure max : 1.90e+10 m2 s
Number of total bins : 768000
Number of fit bins : 691680
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
Accessing contents of a dataset#
To further explore the contents of a Dataset, you can use
e.g. info_dict()
For a quick info, use
print(dataset_cta.info_dict())
{'name': 'dataset-cta', 'counts': 104317, 'excess': 12809.3046875, 'sqrt_ts': 41.41009347393684, 'background': 91507.6953125, 'npred': 91507.68628538586, 'npred_background': 91507.6953125, 'npred_signal': nan, 'exposure_min': <Quantity 62842028. m2 s>, 'exposure_max': <Quantity 1.90242058e+10 m2 s>, 'livetime': <Quantity 5292.00010298 s>, 'ontime': <Quantity 5400. s>, 'counts_rate': <Quantity 19.71220672 1 / s>, 'background_rate': <Quantity 17.29170324 1 / s>, 'excess_rate': <Quantity 2.42050348 1 / s>, 'n_bins': 768000, 'n_fit_bins': 691680, 'stat_type': 'cash', 'stat_sum': nan}
For a quick view, use
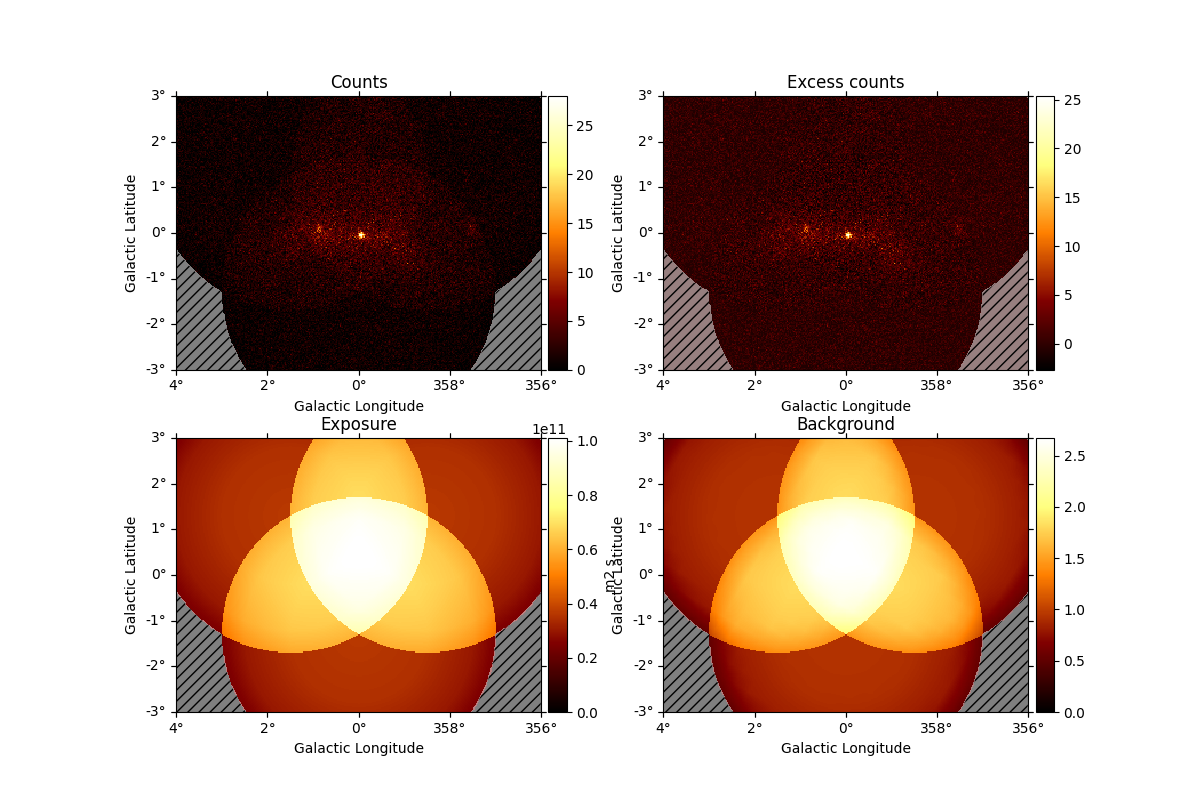
And access individual maps like:
plt.figure()
counts_image = dataset_cta.counts.sum_over_axes()
counts_image.smooth("0.1 deg").plot()
plt.show()
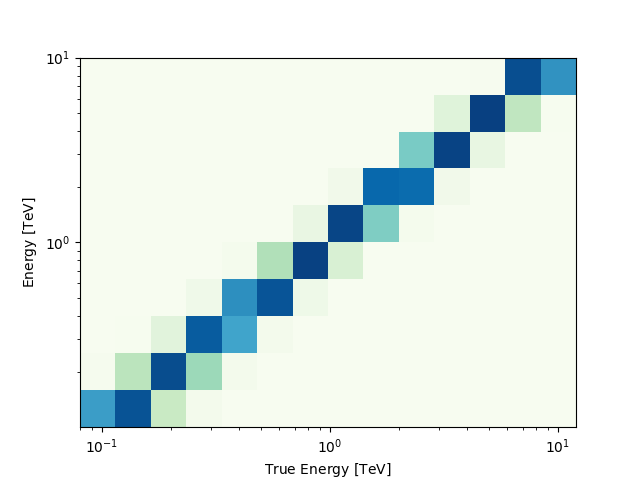
Of course you can also access IRF related maps, e.g. the psf as
PSFMap:
print(dataset_cta.psf)
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'rad', 'energy_true']
shape : (16, 12, 66, 14)
ndim : 4
unit : 1 / sr
dtype : >f4
And use any method on the PSFMap object:
radius = dataset_cta.psf.containment_radius(energy_true=1 * u.TeV, fraction=0.95)
print(radius)
[0.08675] deg
The MapDataset typically also contains the information on the
residual hadronic background, stored in background as a
map:
print(dataset_cta.background)
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (320, 240, 10)
ndim : 3
unit :
dtype : >f4
As a next step we define a minimal model on the dataset using the
models setter:
model = SkyModel.create("pl", "point", name="gc")
model.spatial_model.position = SkyCoord("0d", "0d", frame="galactic")
model_bkg = FoVBackgroundModel(dataset_name="dataset-cta")
dataset_cta.models = [model, model_bkg]
Assigning models to datasets is covered in more detail in Modelling. Printing the dataset will now show the model components:
print(dataset_cta)
MapDataset
----------
Name : dataset-cta
Total counts : 104317
Total background counts : 91507.70
Total excess counts : 12809.31
Predicted counts : 91719.65
Predicted background counts : 91507.69
Predicted excess counts : 211.96
Exposure min : 6.28e+07 m2 s
Exposure max : 1.90e+10 m2 s
Number of total bins : 768000
Number of fit bins : 691680
Fit statistic type : cash
Fit statistic value (-2 log(L)) : 424649.33
Number of models : 2
Number of parameters : 8
Number of free parameters : 5
Component 0: SkyModel
Name : gc
Datasets names : None
Spectral model type : PowerLawSpectralModel
Spatial model type : PointSpatialModel
Temporal model type :
Parameters:
index : 2.000 +/- 0.00
amplitude : 1.00e-12 +/- 0.0e+00 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
lon_0 : 4.650 +/- 0.00 rad
lat_0 : -0.505 +/- 0.00 rad
Component 1: FoVBackgroundModel
Name : dataset-cta-bkg
Datasets names : ['dataset-cta']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
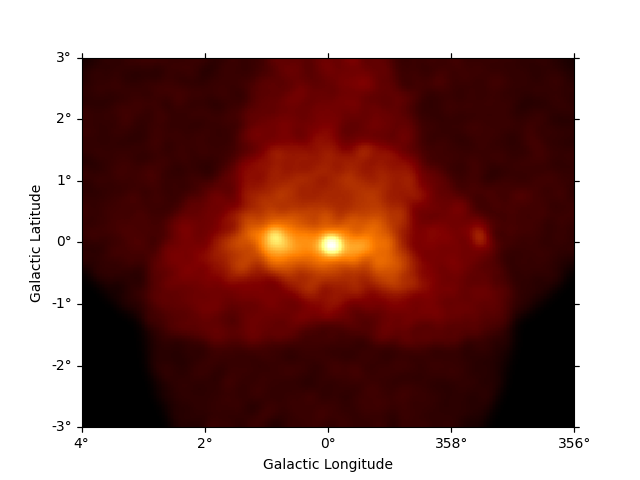
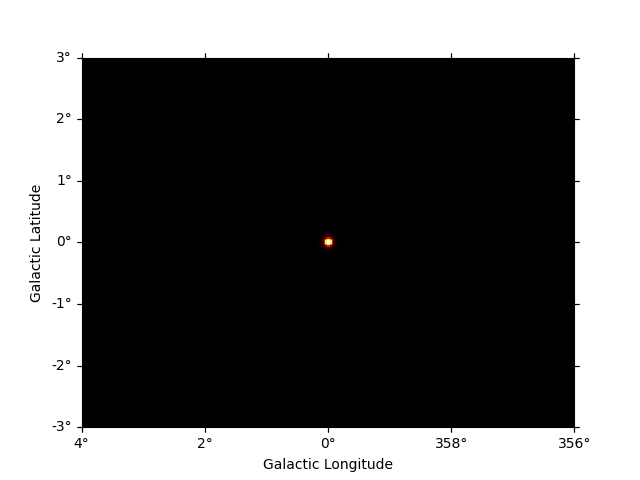
Now we can use the npred method to get a map of the total predicted counts
of the model:
npred = dataset_cta.npred()
npred.sum_over_axes().plot()
plt.show()
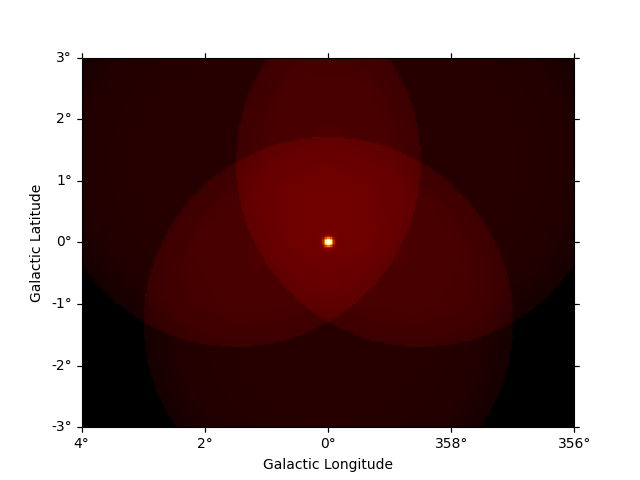
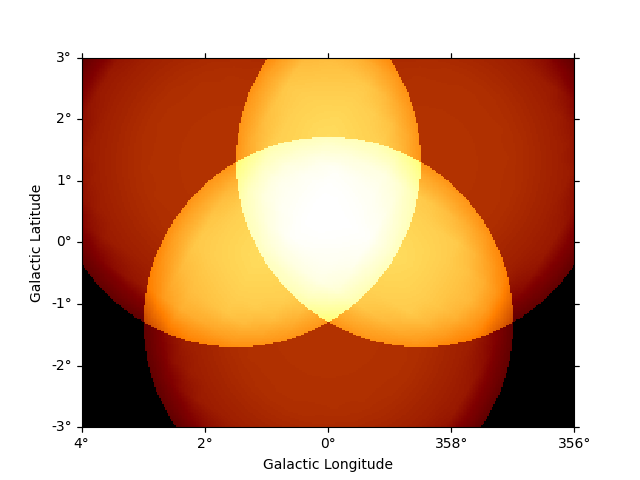
To get the predicted counts from an individual model component we can use:
npred_source = dataset_cta.npred_signal(model_names=["gc"])
npred_source.sum_over_axes().plot()
plt.show()
background contains the background map computed from the
IRF. Internally it will be combined with a FoVBackgroundModel, to
allow for adjusting the background model during a fit. To get the model
corrected background, one can use npred_background.
Using masks#
There are two masks that can be set on a MapDataset, mask_safe and
mask_fit.
The
mask_safeis computed during the data reduction process according to the specified selection cuts, and should not be changed by the user.During modelling and fitting, the user might want to additionally ignore some parts of a reduced dataset, e.g. to restrict the fit to a specific energy range or to ignore parts of the region of interest. This should be done by applying the
mask_fit. To see details of applying masks, please refer to Creating a mask for fitting.
Both the mask_fit and mask_safe must
have the same geom as the counts and
background maps.
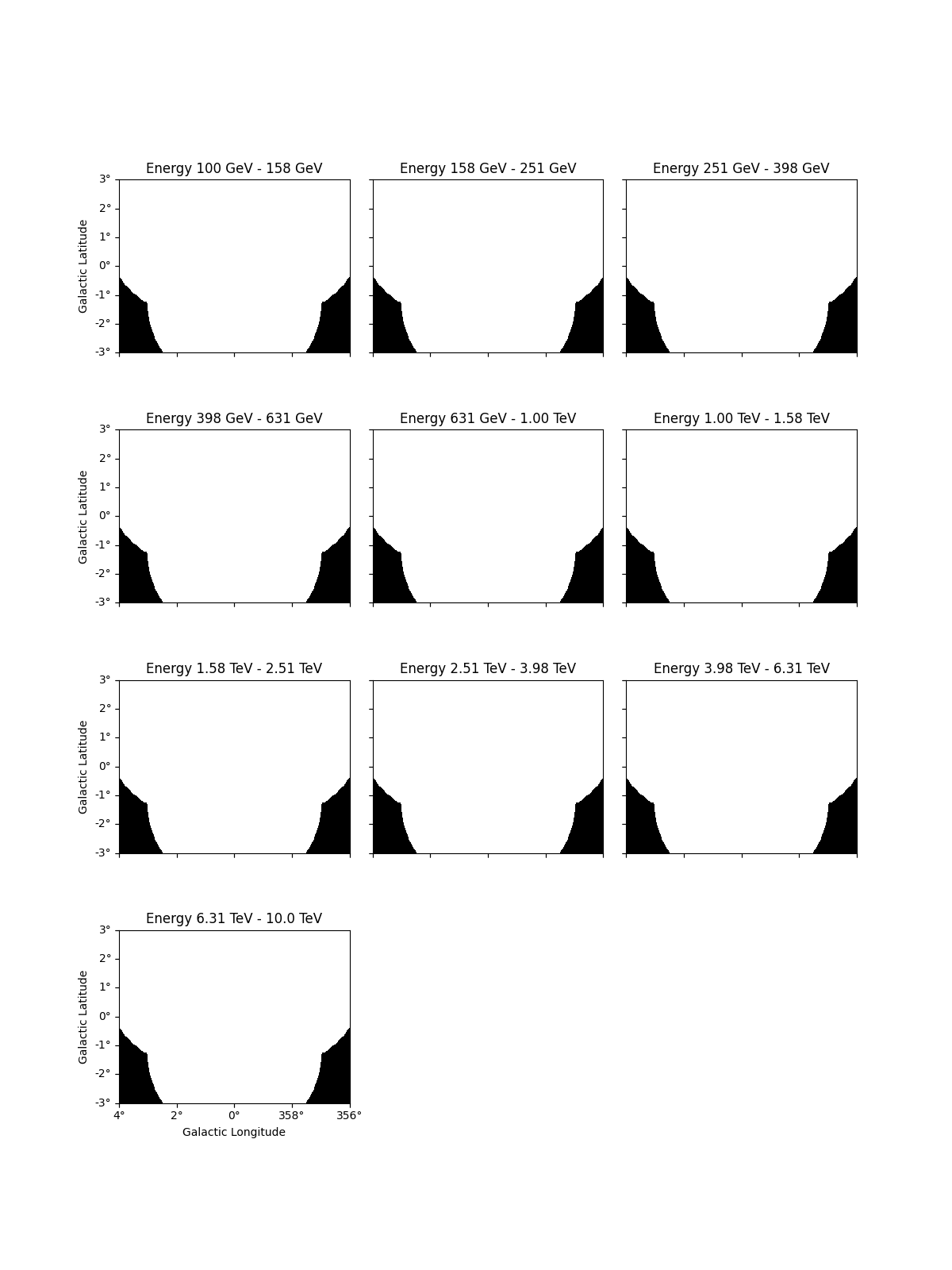
# eg: to see the safe data range
dataset_cta.mask_safe.plot_grid()
plt.show()
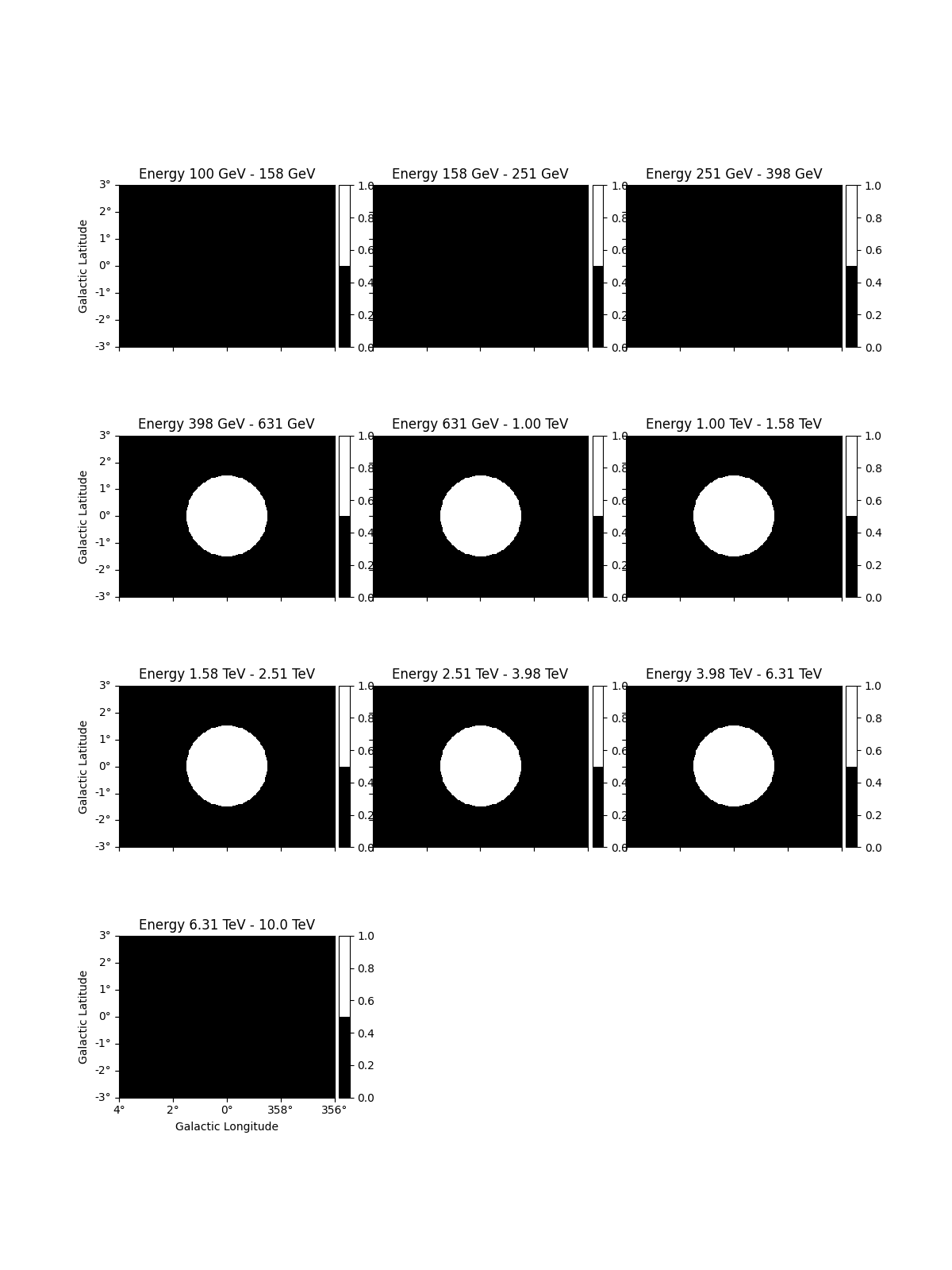
In addition it is possible to define a custom mask_fit:
# To apply a mask fit - in energy and space
region = CircleSkyRegion(SkyCoord("0d", "0d", frame="galactic"), 1.5 * u.deg)
geom = dataset_cta.counts.geom
mask_space = geom.region_mask([region])
mask_energy = geom.energy_mask(0.3 * u.TeV, 8 * u.TeV)
dataset_cta.mask_fit = mask_space & mask_energy
dataset_cta.mask_fit.plot_grid(vmin=0, vmax=1, add_cbar=True)
plt.show()
To access the energy range defined by the mask you can use:
-energy_range_safe : energy range defined by the mask_safe
- energy_range_fit : energy range defined by the mask_fit
- energy_range : the final energy range used in likelihood computation
These methods return two maps, with the min and max energy
values at each spatial pixel
e_min, e_max = dataset_cta.energy_range
# To see the low energy threshold at each point
e_min.plot(add_cbar=True)
plt.show()
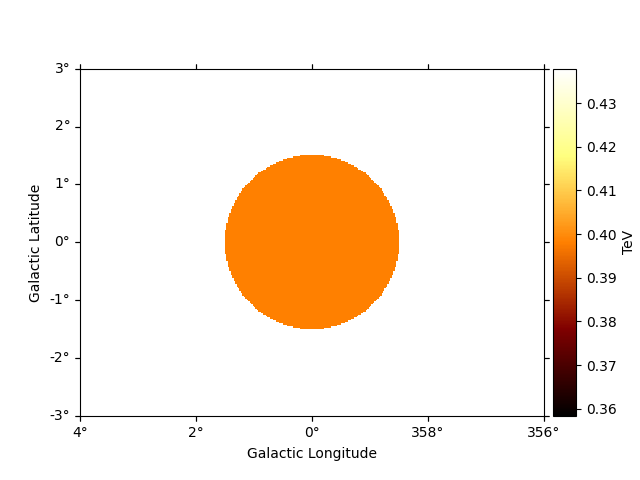
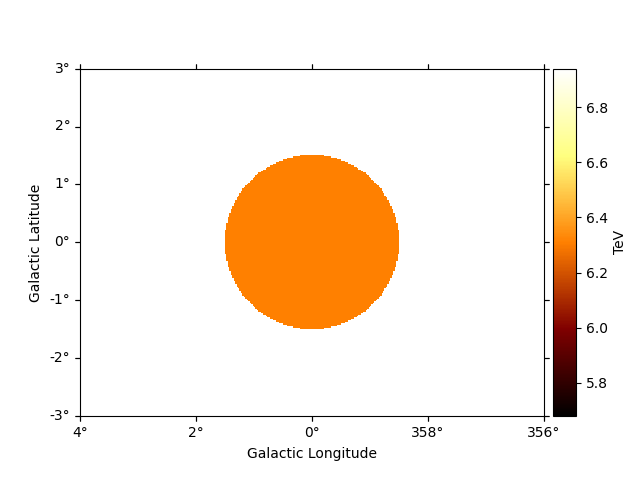
To see the high energy threshold at each point
e_max.plot(add_cbar=True)
plt.show()
Just as for Map objects it is possible to cutout a whole
MapDataset, which will perform the cutout for all maps in
parallel.Optionally one can provide a new name to the resulting dataset:
cutout = dataset_cta.cutout(
position=SkyCoord("0d", "0d", frame="galactic"),
width=2 * u.deg,
name="cta-cutout",
)
cutout.counts.sum_over_axes().plot()
plt.show()
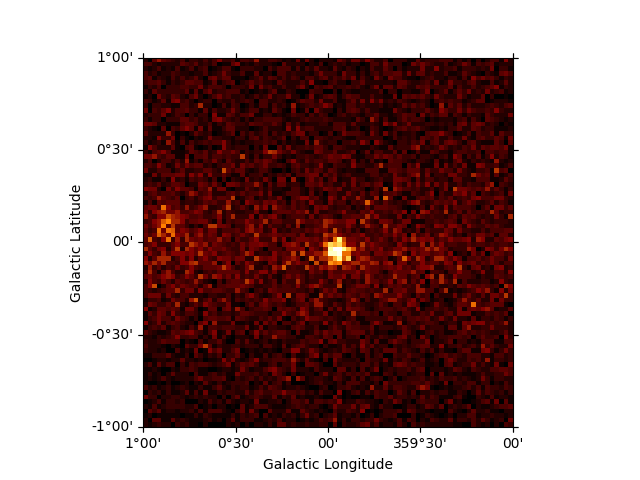
It is also possible to slice a MapDataset in energy:
sliced = dataset_cta.slice_by_energy(
energy_min=1 * u.TeV, energy_max=5 * u.TeV, name="slice-energy"
)
sliced.counts.plot_grid()
plt.show()
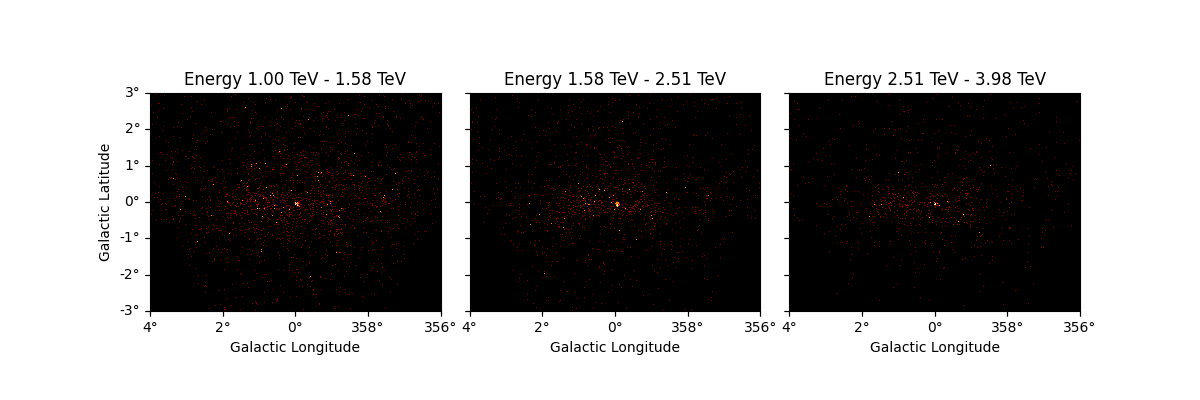
The same operation will be applied to all other maps contained in the
datasets such as mask_fit:
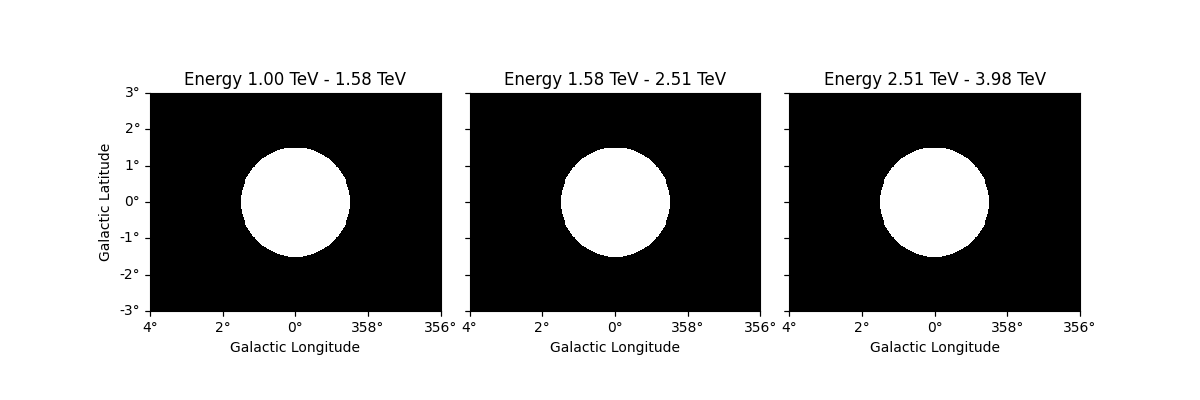
Resampling datasets#
It can often be useful to coarsely rebin an initially computed datasets by a specified factor. This can be done in either spatial or energy axes:
plt.figure()
downsampled = dataset_cta.downsample(factor=8)
downsampled.counts.sum_over_axes().plot()
plt.show()
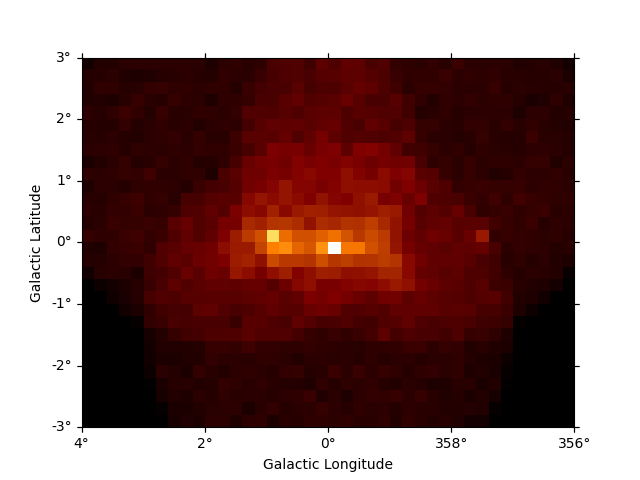
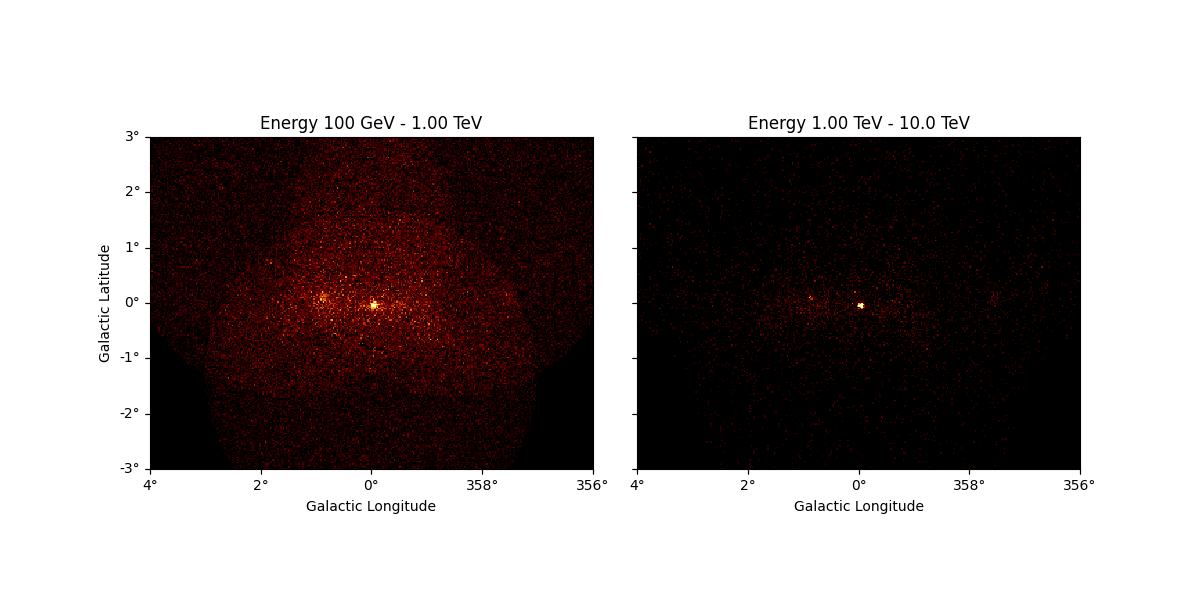
And the same down-sampling process is possible along the energy axis:
downsampled_energy = dataset_cta.downsample(
factor=5, axis_name="energy", name="downsampled-energy"
)
downsampled_energy.counts.plot_grid()
plt.show()
In the printout one can see that the actual number of counts is preserved during the down-sampling:
print(downsampled_energy, dataset_cta)
MapDataset
----------
Name : downsampled-energy
Total counts : 104317
Total background counts : 91507.69
Total excess counts : 12809.31
Predicted counts : 91507.69
Predicted background counts : 91507.69
Predicted excess counts : nan
Exposure min : 6.28e+07 m2 s
Exposure max : 1.90e+10 m2 s
Number of total bins : 153600
Number of fit bins : 22608
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
MapDataset
----------
Name : dataset-cta
Total counts : 104317
Total background counts : 91507.70
Total excess counts : 12809.31
Predicted counts : 91719.65
Predicted background counts : 91507.69
Predicted excess counts : 211.96
Exposure min : 6.28e+07 m2 s
Exposure max : 1.90e+10 m2 s
Number of total bins : 768000
Number of fit bins : 67824
Fit statistic type : cash
Fit statistic value (-2 log(L)) : 44318.32
Number of models : 2
Number of parameters : 8
Number of free parameters : 5
Component 0: SkyModel
Name : gc
Datasets names : None
Spectral model type : PowerLawSpectralModel
Spatial model type : PointSpatialModel
Temporal model type :
Parameters:
index : 2.000 +/- 0.00
amplitude : 1.00e-12 +/- 0.0e+00 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
lon_0 : 4.650 +/- 0.00 rad
lat_0 : -0.505 +/- 0.00 rad
Component 1: FoVBackgroundModel
Name : dataset-cta-bkg
Datasets names : ['dataset-cta']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen): 0.000
reference (frozen): 1.000 TeV
We can also resample the finer binned datasets to an arbitrary coarser energy binning using:
energy_axis_new = MapAxis.from_energy_edges([0.1, 0.3, 1, 3, 10] * u.TeV)
resampled = dataset_cta.resample_energy_axis(energy_axis=energy_axis_new)
resampled.counts.plot_grid(ncols=2)
plt.show()
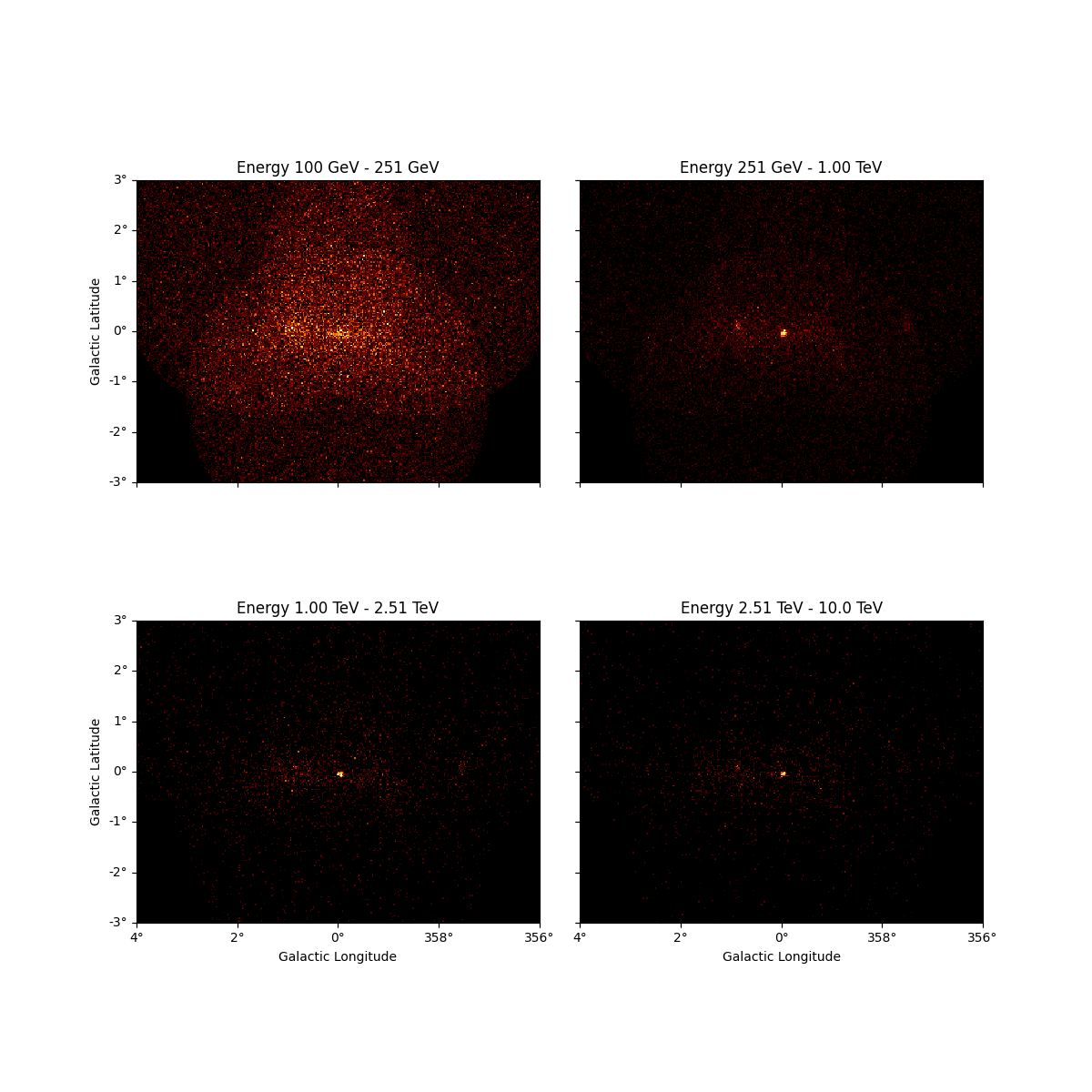
To squash the whole dataset into a single energy bin there is the
to_image() convenience method:

SpectrumDataset#
SpectrumDataset inherits from a MapDataset, and is specially
adapted for 1D spectral analysis, and uses a RegionGeom instead of a
WcsGeom. A MapDataset can be converted to a SpectrumDataset,
by summing the counts and background inside the on_region,
which can then be used for classical spectral analysis. Containment
correction is feasible only for circular regions.
region = CircleSkyRegion(SkyCoord(0, 0, unit="deg", frame="galactic"), 0.5 * u.deg)
spectrum_dataset = dataset_cta.to_spectrum_dataset(
region, containment_correction=True, name="spectrum-dataset"
)
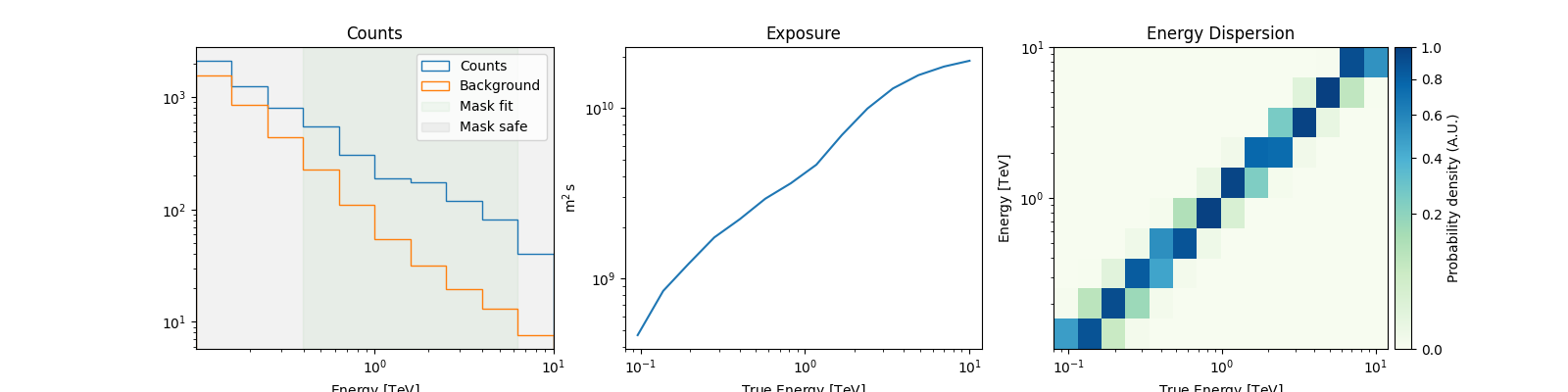
# For a quick look
spectrum_dataset.peek()
plt.show()
A MapDataset can also be integrated over the on_region to create
a MapDataset with a RegionGeom. Complex regions can be handled
and since the full IRFs are used, containment correction is not
required.
reg_dataset = dataset_cta.to_region_map_dataset(region, name="region-map-dataset")
print(reg_dataset)
MapDataset
----------
Name : region-map-dataset
Total counts : 5644
Total background counts : 3323.66
Total excess counts : 2320.34
Predicted counts : 3323.66
Predicted background counts : 3323.66
Predicted excess counts : nan
Exposure min : 4.66e+08 m2 s
Exposure max : 1.89e+10 m2 s
Number of total bins : 10
Number of fit bins : 6
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
FluxPointsDataset#
FluxPointsDataset is a Dataset container for precomputed flux
points, which can be then used in fitting.
flux_points = FluxPoints.read(
"$GAMMAPY_DATA/tests/spectrum/flux_points/diff_flux_points.fits"
)
model = SkyModel(spectral_model=PowerLawSpectralModel(index=2.3))
fp_dataset = FluxPointsDataset(data=flux_points, models=model)
fp_dataset.plot_spectrum()
plt.show()
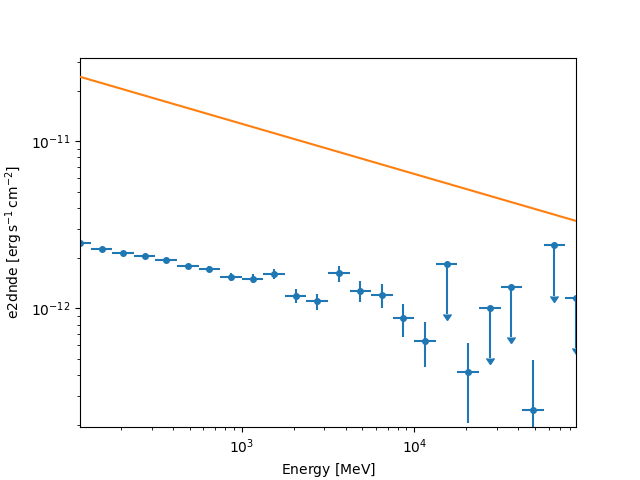
The masks on FluxPointsDataset are ndarray objects
and the data is a
FluxPoints object. The mask_safe,
by default, masks the upper limit points.
print(fp_dataset.mask_safe) # Note: the mask here is simply a numpy array
[[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[ True]]
[[False]]
[[ True]]
[[False]]
[[False]]
[[ True]]
[[False]]
[[False]]]
print(fp_dataset.data) # is a `FluxPoints` object
FluxPoints
----------
geom : RegionGeom
axes : ['lon', 'lat', 'energy']
shape : (1, 1, 24)
quantities : ['norm', 'norm_err', 'norm_ul']
ref. model : pl
n_sigma : 1
n_sigma_ul : 2.0
sqrt_ts_threshold_ul : 2
sed type init : dnde
print(fp_dataset.data_shape()) # number of data points
(24,)
For an example of fitting FluxPoints, see Flux point fitting,
and for using source catalogs see Source catalogs
Datasets#
Datasets are a collection of Dataset objects. They can be of the
same type, or of different types, eg: mix of FluxPointsDataset,
MapDataset and SpectrumDataset.
For modelling and fitting of a list of Dataset objects, you can
either:
(a) Do a joint fitting of all the datasets together OR
(b) Stack the datasets together, and then fit them.
Datasets is a convenient tool to handle joint fitting of
simultaneous datasets. As an example, please see Multi instrument joint 3D and 1D analysis
To see how stacking is performed, please see Stacking Multiple Datasets.
To create a Datasets object, pass a list of Dataset on init, eg
datasets = Datasets([dataset_empty, dataset_cta])
print(datasets)
Datasets
--------
Dataset 0:
Type : MapDataset
Name : dataset-empty
Instrument :
Models :
Dataset 1:
Type : MapDataset
Name : dataset-cta
Instrument :
Models : ['gc', 'dataset-cta-bkg']
If all the datasets have the same type we can also print an info table,
collecting all the information from the individual calls to
info_dict():
display(datasets.info_table()) # quick info of all datasets
print(datasets.names) # unique name of each dataset
name counts excess ... stat_type stat_sum
...
------------- ------ ------------------ ... --------- ------------------
dataset-empty 0 0.0 ... cash nan
dataset-cta 104317 12809.313714614138 ... cash 44318.324424288934
['dataset-empty', 'dataset-cta']
We can access individual datasets in Datasets object by name:
print(datasets["dataset-empty"]) # extracts the first dataset
MapDataset
----------
Name : dataset-empty
Total counts : 0
Total background counts : 0.00
Total excess counts : 0.00
Predicted counts : 0.00
Predicted background counts : 0.00
Predicted excess counts : nan
Exposure min : 0.00e+00 m2 s
Exposure max : 0.00e+00 m2 s
Number of total bins : 110000
Number of fit bins : 0
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
Or by index:
print(datasets[0])
MapDataset
----------
Name : dataset-empty
Total counts : 0
Total background counts : 0.00
Total excess counts : 0.00
Predicted counts : 0.00
Predicted background counts : 0.00
Predicted excess counts : nan
Exposure min : 0.00e+00 m2 s
Exposure max : 0.00e+00 m2 s
Number of total bins : 110000
Number of fit bins : 0
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
Other list type operations work as well such as:
# Use python list convention to remove/add datasets, eg:
datasets.remove("dataset-empty")
print(datasets.names)
['dataset-cta']
Or
['dataset-cta', 'spectrum-dataset']
Let’s create a list of spectrum datasets to illustrate some more functionality:
Datasets
--------
Dataset 0:
Type : SpectrumDatasetOnOff
Name : 23559
Instrument :
Models :
Dataset 1:
Type : SpectrumDatasetOnOff
Name : 23526
Instrument :
Models :
Dataset 2:
Type : SpectrumDatasetOnOff
Name : 23592
Instrument :
Models :
Dataset 3:
Type : SpectrumDatasetOnOff
Name : 23523
Instrument :
Models :
Now we can stack all datasets using stack_reduce():
stacked = datasets.stack_reduce(name="stacked")
print(stacked)
SpectrumDatasetOnOff
--------------------
Name : stacked
Total counts : 459
Total background counts : 27.50
Total excess counts : 431.50
Predicted counts : 45.27
Predicted background counts : 45.27
Predicted excess counts : nan
Exposure min : 2.13e+06 m2 s
Exposure max : 2.63e+09 m2 s
Number of total bins : 80
Number of fit bins : 43
Fit statistic type : wstat
Fit statistic value (-2 log(L)) : 1530.36
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
Total counts_off : 645
Acceptance : 43
Acceptance off : 1149
Or slice all datasets by a given energy range:
datasets_sliced = datasets.slice_by_energy(energy_min="1 TeV", energy_max="10 TeV")
plt.show()
print(datasets_sliced.energy_ranges)
(<Quantity [1.e+09, 1.e+09, 1.e+09, 1.e+09] keV>, <Quantity [1.e+10, 1.e+10, 1.e+10, 1.e+10] keV>)
Total running time of the script: ( 0 minutes 14.326 seconds)