This is a fixed-text formatted version of a Jupyter notebook
You may download all the notebooks as a tar file.
Source files: spectrum_simulation.ipynb | spectrum_simulation.py
1D spectrum simulation#
Prerequisites#
Knowledge of spectral extraction and datasets used in gammapy, see for instance the spectral analysis tutorial
Context#
To simulate a specific observation, it is not always necessary to simulate the full photon list. For many uses cases, simulating directly a reduced binned dataset is enough: the IRFs reduced in the correct geometry are combined with a source model to predict an actual number of counts per bin. The latter is then used to simulate a reduced dataset using Poisson probability distribution.
This can be done to check the feasibility of a measurement, to test whether fitted parameters really provide a good fit to the data etc.
Here we will see how to perform a 1D spectral simulation of a CTA observation, in particular, we will generate OFF observations following the template background stored in the CTA IRFs.
Objective: simulate a number of spectral ON-OFF observations of a source with a power-law spectral model with CTA using the CTA 1DC response, fit them with the assumed spectral model and check that the distribution of fitted parameters is consistent with the input values.
Proposed approach#
We will use the following classes:
Setup#
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
[2]:
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord, Angle
from regions import CircleSkyRegion
from gammapy.datasets import SpectrumDatasetOnOff, SpectrumDataset, Datasets
from gammapy.makers import SpectrumDatasetMaker
from gammapy.modeling import Fit
from gammapy.modeling.models import (
PowerLawSpectralModel,
SkyModel,
)
from gammapy.irf import load_cta_irfs
from gammapy.data import Observation, observatory_locations
from gammapy.maps import MapAxis, RegionGeom
Simulation of a single spectrum#
To do a simulation, we need to define the observational parameters like the livetime, the offset, the assumed integration radius, the energy range to perform the simulation for and the choice of spectral model. We then use an in-memory observation which is convolved with the IRFs to get the predicted number of counts. This is Poission fluctuated using the fake() to get the simulated counts for each observation.
[3]:
# Define simulation parameters parameters
livetime = 1 * u.h
pointing = SkyCoord(0, 0, unit="deg", frame="galactic")
offset = 0.5 * u.deg
# Reconstructed and true energy axis
energy_axis = MapAxis.from_edges(
np.logspace(-0.5, 1.0, 10), unit="TeV", name="energy", interp="log"
)
energy_axis_true = MapAxis.from_edges(
np.logspace(-1.2, 2.0, 31), unit="TeV", name="energy_true", interp="log"
)
on_region_radius = Angle("0.11 deg")
center = pointing.directional_offset_by(
position_angle=0 * u.deg, separation=offset
)
on_region = CircleSkyRegion(center=center, radius=on_region_radius)
[4]:
# Define spectral model - a simple Power Law in this case
model_simu = PowerLawSpectralModel(
index=3.0,
amplitude=2.5e-12 * u.Unit("cm-2 s-1 TeV-1"),
reference=1 * u.TeV,
)
print(model_simu)
# we set the sky model used in the dataset
model = SkyModel(spectral_model=model_simu, name="source")
PowerLawSpectralModel
type name value unit error min max frozen is_norm link
-------- --------- ---------- -------------- --------- --- --- ------ ------- ----
spectral index 3.0000e+00 0.000e+00 nan nan False False
spectral amplitude 2.5000e-12 cm-2 s-1 TeV-1 0.000e+00 nan nan False True
spectral reference 1.0000e+00 TeV 0.000e+00 nan nan True False
[5]:
# Load the IRFs
# In this simulation, we use the CTA-1DC irfs shipped with gammapy.
irfs = load_cta_irfs(
"$GAMMAPY_DATA/cta-1dc/caldb/data/cta/1dc/bcf/South_z20_50h/irf_file.fits"
)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
[6]:
location = observatory_locations["cta_south"]
obs = Observation.create(
pointing=pointing,
livetime=livetime,
irfs=irfs,
location=location,
)
print(obs)
Observation
obs id : 0
tstart : 51544.00
tstop : 51544.04
duration : 3600.00 s
pointing (icrs) : 266.4 deg, -28.9 deg
deadtime fraction : 0.0%
[7]:
# Make the SpectrumDataset
geom = RegionGeom.create(region=on_region, axes=[energy_axis])
dataset_empty = SpectrumDataset.create(
geom=geom, energy_axis_true=energy_axis_true, name="obs-0"
)
maker = SpectrumDatasetMaker(selection=["exposure", "edisp", "background"])
dataset = maker.run(dataset_empty, obs)
[8]:
# Set the model on the dataset, and fake
dataset.models = model
dataset.fake(random_state=42)
print(dataset)
SpectrumDataset
---------------
Name : obs-0
Total counts : 298
Total background counts : 22.29
Total excess counts : 275.71
Predicted counts : 303.66
Predicted background counts : 22.29
Predicted excess counts : 281.37
Exposure min : 2.53e+08 m2 s
Exposure max : 1.77e+10 m2 s
Number of total bins : 9
Number of fit bins : 9
Fit statistic type : cash
Fit statistic value (-2 log(L)) : -1811.58
Number of models : 1
Number of parameters : 3
Number of free parameters : 2
Component 0: SkyModel
Name : source
Datasets names : None
Spectral model type : PowerLawSpectralModel
Spatial model type :
Temporal model type :
Parameters:
index : 3.000 +/- 0.00
amplitude : 2.50e-12 +/- 0.0e+00 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
You can see that background counts are now simulated
On-Off analysis#
To do an on off spectral analysis, which is the usual science case, the standard would be to use SpectrumDatasetOnOff, which uses the acceptance to fake off-counts
[9]:
dataset_on_off = SpectrumDatasetOnOff.from_spectrum_dataset(
dataset=dataset, acceptance=1, acceptance_off=5
)
dataset_on_off.fake(npred_background=dataset.npred_background())
print(dataset_on_off)
SpectrumDatasetOnOff
--------------------
Name : HEEdFEpA
Total counts : 314
Total background counts : 22.40
Total excess counts : 291.60
Predicted counts : 303.82
Predicted background counts : 22.45
Predicted excess counts : 281.37
Exposure min : 2.53e+08 m2 s
Exposure max : 1.77e+10 m2 s
Number of total bins : 9
Number of fit bins : 9
Fit statistic type : wstat
Fit statistic value (-2 log(L)) : 10.32
Number of models : 1
Number of parameters : 3
Number of free parameters : 2
Component 0: SkyModel
Name : source
Datasets names : None
Spectral model type : PowerLawSpectralModel
Spatial model type :
Temporal model type :
Parameters:
index : 3.000 +/- 0.00
amplitude : 2.50e-12 +/- 0.0e+00 1 / (cm2 s TeV)
reference (frozen): 1.000 TeV
Total counts_off : 112
Acceptance : 9
Acceptance off : 45
You can see that off counts are now simulated as well. We now simulate several spectra using the same set of observation conditions.
[10]:
%%time
n_obs = 100
datasets = Datasets()
for idx in range(n_obs):
dataset_on_off.fake(
random_state=idx, npred_background=dataset.npred_background()
)
dataset_fake = dataset_on_off.copy(name=f"obs-{idx}")
dataset_fake.meta_table["OBS_ID"] = [idx]
datasets.append(dataset_fake)
CPU times: user 2.36 s, sys: 43.2 ms, total: 2.41 s
Wall time: 2.67 s
[11]:
table = datasets.info_table()
table
[11]:
| name | counts | excess | sqrt_ts | background | npred | npred_background | npred_signal | exposure_min | exposure_max | livetime | ontime | counts_rate | background_rate | excess_rate | n_bins | n_fit_bins | stat_type | stat_sum | counts_off | acceptance | acceptance_off | alpha |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m2 s | m2 s | s | s | 1 / s | 1 / s | 1 / s | ||||||||||||||||
| str6 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | int64 | int64 | str5 | float64 | int64 | float64 | float64 | float64 |
| obs-0 | 317 | 298.6000061035156 | 27.08240180813126 | 18.399999618530273 | 68.16666751313541 | 68.16666751313541 | nan | 252718176.0 | 17719697408.0 | 3600.000000000002 | 3600.000000000002 | 0.08805555555555551 | 0.0051111110051472956 | 0.08294444613986542 | 9 | 9 | wstat | 738.7245834271496 | 92 | 9.0 | 45.0 | 0.20000000298023224 |
| obs-1 | 275 | 253.0 | 23.76785365487285 | 22.0 | 64.16666666666669 | 64.16666666666669 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07638888888888885 | 0.006111111111111108 | 0.07027777777777774 | 9 | 9 | wstat | 575.779512784738 | 110 | 9.0 | 45.0 | 0.2 |
| obs-2 | 293 | 272.4 | 25.17110555404655 | 20.6 | 66.0 | 66.0 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08138888888888884 | 0.00572222222222222 | 0.07566666666666662 | 9 | 9 | wstat | 645.4993824075303 | 103 | 9.0 | 45.0 | 0.2 |
| obs-3 | 280 | 257.6 | 23.982951737405354 | 22.4 | 65.33333333333334 | 65.33333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07777777777777774 | 0.006222222222222218 | 0.07155555555555553 | 9 | 9 | wstat | 585.9241546985872 | 112 | 9.0 | 45.00000000000001 | 0.19999999999999998 |
| obs-4 | 337 | 316.4 | 27.682709945184747 | 20.6 | 73.33333333333334 | 73.33333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.09361111111111106 | 0.00572222222222222 | 0.08788888888888884 | 9 | 9 | wstat | 787.314723949448 | 103 | 9.0 | 45.0 | 0.2 |
| obs-5 | 283 | 258.6 | 23.727154782347895 | 24.400000000000002 | 67.50000000000001 | 67.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07861111111111108 | 0.006777777777777775 | 0.0718333333333333 | 9 | 9 | wstat | 574.8525737727499 | 122 | 9.0 | 45.0 | 0.2 |
| obs-6 | 330 | 307.6 | 26.889184475727866 | 22.400000000000006 | 73.66666666666667 | 73.66666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.09166666666666662 | 0.006222222222222221 | 0.0854444444444444 | 9 | 9 | wstat | 734.2414755285937 | 112 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-7 | 283 | 257.2 | 23.43087974795853 | 25.8 | 68.66666666666667 | 68.66666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07861111111111108 | 0.007166666666666663 | 0.07144444444444441 | 9 | 9 | wstat | 552.3698779448044 | 129 | 9.0 | 45.0 | 0.2 |
| obs-8 | 308 | 284.6 | 25.42049273328331 | 23.400000000000002 | 70.83333333333334 | 70.83333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08555555555555551 | 0.006499999999999997 | 0.07905555555555552 | 9 | 9 | wstat | 652.2621572567633 | 117 | 9.0 | 45.0 | 0.2 |
| obs-9 | 299 | 278.6 | 25.57085071486863 | 20.4 | 66.83333333333334 | 66.83333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08305555555555551 | 0.005666666666666664 | 0.07738888888888885 | 9 | 9 | wstat | 666.9062670260786 | 102 | 9.0 | 45.00000000000001 | 0.19999999999999998 |
| obs-10 | 310 | 294.8 | 27.488972161356774 | 15.2 | 64.33333333333334 | 64.33333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08611111111111107 | 0.00422222222222222 | 0.08188888888888884 | 9 | 9 | wstat | 768.5404492529331 | 76 | 9.0 | 45.00000000000001 | 0.19999999999999998 |
| obs-11 | 285 | 261.0 | 23.933833745454685 | 24.000000000000004 | 67.50000000000001 | 67.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07916666666666662 | 0.0066666666666666645 | 0.07249999999999997 | 9 | 9 | wstat | 583.448753292733 | 120 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-12 | 299 | 278.0 | 25.4325263895117 | 21.000000000000004 | 67.33333333333336 | 67.33333333333336 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08305555555555551 | 0.005833333333333331 | 0.07722222222222218 | 9 | 9 | wstat | 655.2768246480357 | 105 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-13 | 309 | 287.4 | 25.877406235012618 | 21.6 | 69.50000000000001 | 69.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08583333333333329 | 0.0059999999999999975 | 0.07983333333333328 | 9 | 9 | wstat | 672.355748274336 | 108 | 9.0 | 45.0 | 0.2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| obs-85 | 297 | 274.6 | 24.998935845735573 | 22.4 | 68.16666666666667 | 68.16666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08249999999999996 | 0.006222222222222218 | 0.07627777777777775 | 9 | 9 | wstat | 631.6298361198053 | 112 | 9.0 | 45.00000000000001 | 0.19999999999999998 |
| obs-86 | 286 | 268.2 | 25.42340896951546 | 17.800000000000004 | 62.50000000000001 | 62.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.0794444444444444 | 0.004944444444444443 | 0.07449999999999996 | 9 | 9 | wstat | 655.2082535886425 | 89 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-87 | 333 | 313.2 | 27.646851451147217 | 19.8 | 72.00000000000001 | 72.00000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.09249999999999996 | 0.005499999999999997 | 0.08699999999999995 | 9 | 9 | wstat | 770.1313412321031 | 99 | 9.0 | 45.0 | 0.2 |
| obs-88 | 315 | 296.8 | 27.0172059251097 | 18.200000000000003 | 67.66666666666669 | 67.66666666666669 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08749999999999995 | 0.005055555555555554 | 0.0824444444444444 | 9 | 9 | wstat | 734.6476098262428 | 91 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-89 | 287 | 265.0 | 24.49456558680515 | 22.000000000000004 | 66.16666666666667 | 66.16666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07972222222222218 | 0.006111111111111109 | 0.07361111111111107 | 9 | 9 | wstat | 611.2334063685089 | 110 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-90 | 286 | 267.0 | 25.131221887043438 | 19.000000000000004 | 63.50000000000001 | 63.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.0794444444444444 | 0.005277777777777776 | 0.07416666666666663 | 9 | 9 | wstat | 635.8304116110406 | 95 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-91 | 285 | 259.8 | 23.67754591931069 | 25.200000000000003 | 68.50000000000001 | 68.50000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.07916666666666662 | 0.0069999999999999975 | 0.07216666666666663 | 9 | 9 | wstat | 573.3564742963381 | 126 | 9.0 | 45.0 | 0.2 |
| obs-92 | 313 | 289.4 | 25.664935420194176 | 23.6 | 71.83333333333336 | 71.83333333333336 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.0869444444444444 | 0.006555555555555552 | 0.08038888888888884 | 9 | 9 | wstat | 669.4496142825196 | 118 | 9.0 | 45.0 | 0.2 |
| obs-93 | 302 | 283.2 | 26.123867522605497 | 18.8 | 66.00000000000001 | 66.00000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08388888888888885 | 0.00522222222222222 | 0.07866666666666662 | 9 | 9 | wstat | 700.5066738339824 | 94 | 9.0 | 45.0 | 0.2 |
| obs-94 | 322 | 300.2 | 26.574029757473202 | 21.8 | 71.83333333333334 | 71.83333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.0894444444444444 | 0.006055555555555553 | 0.08338888888888885 | 9 | 9 | wstat | 715.2570498037147 | 109 | 9.0 | 45.0 | 0.2 |
| obs-95 | 305 | 280.6 | 25.030731260493152 | 24.400000000000006 | 71.16666666666667 | 71.16666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08472222222222218 | 0.006777777777777776 | 0.07794444444444441 | 9 | 9 | wstat | 643.2846879404793 | 122 | 9.0 | 44.99999999999999 | 0.20000000000000004 |
| obs-96 | 301 | 277.4 | 24.969845969421428 | 23.6 | 69.83333333333334 | 69.83333333333334 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08361111111111107 | 0.006555555555555552 | 0.07705555555555552 | 9 | 9 | wstat | 626.7396842428624 | 118 | 9.0 | 45.0 | 0.2 |
| obs-97 | 290 | 271.2 | 25.417982194454826 | 18.8 | 64.00000000000001 | 64.00000000000001 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08055555555555552 | 0.00522222222222222 | 0.0753333333333333 | 9 | 9 | wstat | 650.9762484493856 | 94 | 9.0 | 45.0 | 0.2 |
| obs-98 | 301 | 280.6 | 25.687832964675007 | 20.400000000000002 | 67.16666666666667 | 67.16666666666667 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08361111111111107 | 0.0056666666666666645 | 0.07794444444444441 | 9 | 9 | wstat | 664.5620099404952 | 102 | 9.0 | 45.0 | 0.2 |
| obs-99 | 323 | 302.2 | 26.85707842376871 | 20.8 | 71.16666666666669 | 71.16666666666669 | nan | 252718170.97287694 | 17719697919.59929 | 3600.000000000002 | 3600.000000000002 | 0.08972222222222218 | 0.005777777777777775 | 0.0839444444444444 | 9 | 9 | wstat | 733.1576598768017 | 104 | 9.0 | 45.0 | 0.2 |
Before moving on to the fit let’s have a look at the simulated observations.
[12]:
fix, axes = plt.subplots(1, 3, figsize=(12, 4))
axes[0].hist(table["counts"])
axes[0].set_xlabel("Counts")
axes[1].hist(table["counts_off"])
axes[1].set_xlabel("Counts Off")
axes[2].hist(table["excess"])
axes[2].set_xlabel("excess");
Now, we fit each simulated spectrum individually
[13]:
%%time
results = []
fit = Fit()
for dataset in datasets:
dataset.models = model.copy()
result = fit.optimize(dataset)
results.append(
{
"index": result.parameters["index"].value,
"amplitude": result.parameters["amplitude"].value,
}
)
CPU times: user 40.7 s, sys: 247 ms, total: 40.9 s
Wall time: 45.1 s
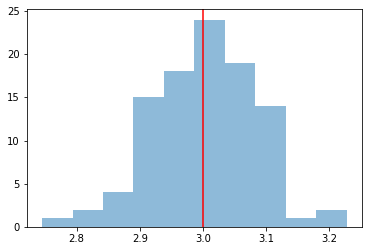
We take a look at the distribution of the fitted indices. This matches very well with the spectrum that we initially injected.
[14]:
index = np.array([_["index"] for _ in results])
plt.hist(index, bins=10, alpha=0.5)
plt.axvline(x=model_simu.parameters["index"].value, color="red")
print(f"index: {index.mean()} += {index.std()}")
index: 3.003692555041602 += 0.08081469526978655
Exercises#
Change the observation time to something longer or shorter. Does the observation and spectrum results change as you expected?
Change the spectral model, e.g. add a cutoff at 5 TeV, or put a steep-spectrum source with spectral index of 4.0
Simulate spectra with the spectral model we just defined. How much observation duration do you need to get back the injected parameters?
[ ]:
