This is a fixed-text formatted version of a Jupyter notebook.
- Try online
- You can contribute with your own notebooks in this GitHub repository.
- Source files: nddata_demo.ipynb | nddata_demo.py
How to use the NDDataArray class¶
Introduction¶
This notebook explains how to use the class gammapy.utils.nddata.NDDataArray
The NDDataArray is basically an numpy array with associated axes and convenience methods for interpolation. For now only the scipy RegularGridInterpolator can be used, i.e. available interpolation methods are “nearest neighbour” and “linear”. A spline interpolator will be added in the future. The interpolation behaviour (“log”, “linear”) can be set for each axis individually.
The NDDataArray is currently used in the following classes
- gammapy.irf.EffectiveAreaTable
- gammapy.irf.EffectiveAreaTable2D
- gammapy.irf.EnergyDispersion
- gammapy.spectrum.CountsSpectrum
- Probably some more by now …
Feedback welcome!
Setup¶
As usual, we’ll start with some setup …
In [1]:
%matplotlib inline
import matplotlib.pyplot as plt
In [2]:
from gammapy.utils.nddata import NDDataArray, DataAxis, BinnedDataAxis
from gammapy.utils.energy import Energy, EnergyBounds
import numpy as np
import astropy.units as u
1D example¶
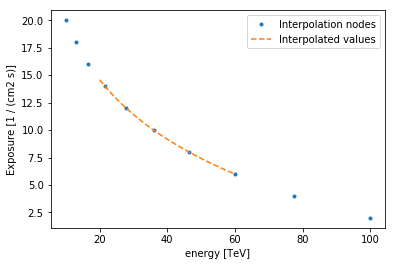
Let’s start with a simple example. A one dimensional array storing an
exposure in cm-2 s-1 as a function of energy. The energy axis is log
spaced and thus also the interpolation shall take place in log.
In [3]:
energies = Energy.equal_log_spacing(10, 100, 10, unit=u.TeV)
x_axis = DataAxis(energies, name="energy", interpolation_mode="log")
data = np.arange(20, 0, -2) / u.cm ** 2 / u.s
nddata = NDDataArray(axes=[x_axis], data=data)
print(nddata)
print(nddata.axis("energy"))
NDDataArray summary info
energy : size = 10, min = 10.000 TeV, max = 100.000 TeV
Data : size = 10, min = 2.000 1 / (cm2 s), max = 20.000 1 / (cm2 s)
DataAxis
Name: energy
Unit: TeV
Nodes: 10
Interpolation mode: log
In [4]:
eval_energies = np.linspace(2, 6, 20) * 1e4 * u.GeV
eval_exposure = nddata.evaluate(energy=eval_energies, method="linear")
plt.plot(
nddata.axis("energy").nodes.value,
nddata.data.value,
".",
label="Interpolation nodes",
)
print(nddata.axis("energy").nodes)
plt.plot(
eval_energies.to("TeV").value,
eval_exposure,
"--",
label="Interpolated values",
)
plt.xlabel("{} [{}]".format(nddata.axes[0].name, nddata.axes[0].unit))
plt.ylabel("{} [{}]".format("Exposure", nddata.data.unit))
plt.legend();
[ 10. 12.91549665 16.68100537 21.5443469 27.82559402
35.93813664 46.41588834 59.94842503 77.42636827 100. ] TeV
2D example¶
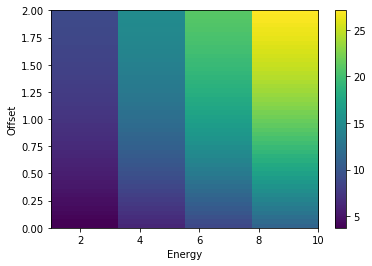
Another common use case is to store a Quantity as a function of field of view offset and energy. The following shows how to use the NDDataArray to slice the data array at any values of offset and energy
In [5]:
energy_data = EnergyBounds.equal_log_spacing(1, 10, 50, unit=u.TeV)
energy_axis = BinnedDataAxis(
lo=energy_data.lower_bounds,
hi=energy_data.upper_bounds,
name="energy",
interpolation_mode="log",
)
offset_data = np.linspace(0, 2, 4) * u.deg
offset_axis = DataAxis(offset_data, name="offset")
data_temp = 10 * np.exp(-energy_data.log_centers.value / 10)
data = np.outer(data_temp, (offset_data.value + 1))
nddata2d = NDDataArray(
axes=[energy_axis, offset_axis], data=data * u.Unit("cm-2 s-1 TeV-1")
)
print(nddata2d)
extent_x = nddata2d.axis("energy").bins[[0, -1]].value
extent_y = nddata2d.axis("offset").nodes[[0, -1]].value
extent = extent_x[0], extent_x[1], extent_y[0], extent_y[1]
plt.imshow(nddata2d.data.value, extent=extent, aspect="auto")
plt.xlabel("Energy")
plt.ylabel("Offset")
plt.colorbar();
NDDataArray summary info
energy : size = 50, min = 1.023 TeV, max = 9.772 TeV
offset : size = 4, min = 0.000 deg, max = 2.000 deg
Data : size = 200, min = 3.763 1 / (cm2 s TeV), max = 27.082 1 / (cm2 s TeV)
In [6]:
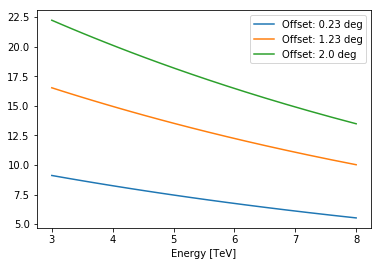
offsets = [0.23, 1.23, 2] * u.deg
eval_energies = Energy.equal_log_spacing(3, 8, 20, u.TeV)
for offset in offsets:
slice_ = nddata2d.evaluate(offset=offset, energy=eval_energies)
plt.plot(eval_energies.value, slice_, label="Offset: {}".format(offset))
plt.xlabel("Energy [TeV]")
plt.legend();
In [7]: