This is a fixed-text formatted version of a Jupyter notebook
You can contribute with your own notebooks in this GitHub repository.
Source files: analysis_mwl.ipynb | analysis_mwl.py
Joint modeling, fitting, and serialization¶
This tutorial illustrates how to perfom a joint modeling and fitting of the Crab Nebula spectrum using different datasets. We look at the gamma-ray emission from the Crab nebula between 10 GeV and 100 TeV. The spectral parameters are optimized by combining a 3D analysis of Fermi-LAT data, a ON/OFF spectral analysis of HESS data, and flux points from HAWC.
In this tutorial we are going to use pre-made datasets. We prepared maps of the Crab region as seen by Fermi-LAT using the same event selection than the 3FHL catalog (7 years of data with energy from 10 GeV to 2 TeV). For the HESS ON/OFF analysis we used two observations from the first public data release with a significant signal from energy of about 600 GeV to 10 TeV. These observations have an offset of 0.5° and a zenith angle of 45-48°. The HAWC flux points data are taken from a recent analysis based on 2.5 years of data with energy between 300 Gev and 300 TeV.
More details on how to prepare datasets with the high and low level interfaces are available in these tutorials:
[1]:
from astropy import units as u
import matplotlib.pyplot as plt
from gammapy.modeling import Fit, Datasets
from gammapy.spectrum import (
FluxPoints,
FluxPointsEstimator,
FluxPointsDataset,
SpectrumDatasetOnOff,
)
from gammapy.maps import MapAxis
from pathlib import Path
Data and models files¶
The datasets serialization produce YAML files listing the datasets and models. In the following cells we show an example containning only the Fermi-LAT dataset and the Crab model.
Fermi-LAT-3FHL_datasets.yaml:
datasets:
- name: Fermi-LAT
type: MapDataset
likelihood: cash
models:
- Crab Nebula
background: background
filename: $GAMMAPY_DATA/fermi-3fhl-crab/Fermi-LAT-3FHL_data_Fermi-LAT.fits
We used as model a point source with a log-parabola spectrum. The initial parameters were taken from the latest Fermi-LAT catalog 4FGL, then we have re-optimized the spectral parameters for our dataset in the 10 GeV - 2 TeV energy range (fixing the source position).
Fermi-LAT-3FHL_models.yaml:
components:
- name: Crab Nebula
type: SkyModel
spatial:
type: PointSpatialModel
frame: icrs
parameters:
- name: lon_0
value: 83.63310241699219
unit: deg
min: .nan
max: .nan
frozen: true
- name: lat_0
value: 22.019899368286133
unit: deg
min: -90.0
max: 90.0
frozen: true
spectral:
type: LogParabolaSpectralModel
parameters:
- name: amplitude
value: 0.3415498620816483
unit: cm-2 s-1 TeV-1
min: .nan
max: .nan
frozen: false
- name: reference
value: 5.054833602905273e-05
unit: TeV
min: .nan
max: .nan
frozen: true
- name: alpha
value: 2.510798031388936
unit: ''
min: .nan
max: .nan
frozen: false
- name: beta
value: -0.022476498188855533
unit: ''
min: .nan
max: .nan
frozen: false
- name: background
type: BackgroundModel
parameters:
- name: norm
value: 0.9544383244743555
unit: ''
min: 0.0
max: .nan
frozen: false
- name: tilt
value: 0.0
unit: ''
min: .nan
max: .nan
frozen: true
- name: reference
value: 1.0
unit: TeV
min: .nan
max: .nan
frozen: true
Reading different datasets¶
Fermi-LAT 3FHL: map dataset for 3D analysis¶
For now we let’s use the datasets serialization only to read the 3D MapDataset associated to Fermi-LAT 3FHL data and models.
[2]:
path = "$GAMMAPY_DATA/fermi-3fhl-crab/Fermi-LAT-3FHL"
filedata = Path(path + "_datasets.yaml")
filemodel = Path(path + "_models.yaml")
datasets = Datasets.read(filedata=filedata, filemodel=filemodel)
dataset_fermi = datasets[0]
We get the Crab model in order to share it with the other datasets
[3]:
crab_model = dataset_fermi.models["Crab Nebula"]
crab_spec = crab_model.spectral_model
print(crab_spec)
LogParabolaSpectralModel
name value error unit min max frozen
--------- ---------- ----- -------------- --- --- ------
amplitude 3.415e-01 nan cm-2 s-1 TeV-1 nan nan False
reference 5.055e-05 nan TeV nan nan True
alpha 2.511e+00 nan nan nan False
beta -2.248e-02 nan nan nan False
HESS-DL3: 1D ON/OFF dataset for spectral fitting¶
The ON/OFF datasets can be read from PHA files following the OGIP standards. We read the PHA files from each observation, and compute a stacked dataset for simplicity. Then the Crab spectral model previously defined is added to the dataset.
[4]:
datasets = []
for obs_id in [23523, 23526]:
dataset = SpectrumDatasetOnOff.from_ogip_files(
f"$GAMMAPY_DATA/joint-crab/spectra/hess/pha_obs{obs_id}.fits"
)
datasets.append(dataset)
dataset_hess = Datasets(datasets).stack_reduce()
dataset_hess.name = "HESS"
dataset_hess.models = crab_model
HAWC: 1D dataset for flux point fitting¶
The HAWC flux point are taken from https://arxiv.org/pdf/1905.12518.pdf. Then these flux points are read from a pre-made FITS file and passed to a FluxPointsDataset together with the source spectral model.
[5]:
# read flux points from https://arxiv.org/pdf/1905.12518.pdf
filename = "$GAMMAPY_DATA/hawc_crab/HAWC19_flux_points.fits"
flux_points_hawc = FluxPoints.read(filename)
dataset_hawc = FluxPointsDataset(crab_model, flux_points_hawc, name="HAWC")
Datasets serialization¶
The datasets object contains each dataset previously defined. It can be saved on disk as datasets.yaml, models.yaml, and several data files specific to each dataset. Then the datasets can be rebuild later from these files.
[6]:
datasets = Datasets([dataset_fermi, dataset_hess, dataset_hawc])
path = Path("crab-3datasets")
path.mkdir(exist_ok=True)
datasets.write(path=path, prefix="crab_10GeV_100TeV", overwrite=True)
filedata = path / "crab_10GeV_100TeV_datasets.yaml"
filemodel = path / "crab_10GeV_100TeV_models.yaml"
datasets = Datasets.read(filedata=filedata, filemodel=filemodel)
Joint analysis¶
We run the fit on the Datasets object that include a dataset for each instrument
[7]:
%%time
fit_joint = Fit(datasets)
results_joint = fit_joint.run()
print(results_joint)
print(results_joint.parameters.to_table())
OptimizeResult
backend : minuit
method : minuit
success : True
message : Optimization terminated successfully.
nfev : 866
total stat : -14820.65
name value error unit min max frozen
--------- --------- --------- -------------- ---------- --------- ------
lon_0 8.363e+01 0.000e+00 deg nan nan True
lat_0 2.202e+01 0.000e+00 deg -9.000e+01 9.000e+01 True
amplitude 3.721e-03 6.347e-04 cm-2 s-1 TeV-1 nan nan False
reference 5.055e-05 0.000e+00 TeV nan nan True
alpha 1.246e+00 3.935e-02 nan nan False
beta 6.247e-02 2.178e-03 nan nan False
norm 9.826e-01 3.029e-01 0.000e+00 nan False
tilt 0.000e+00 0.000e+00 nan nan True
reference 1.000e+00 0.000e+00 TeV nan nan True
CPU times: user 10.6 s, sys: 118 ms, total: 10.7 s
Wall time: 10.8 s
Let’s display only the parameters of the Crab spectral model
[8]:
crab_spec = datasets[0].models["Crab Nebula"].spectral_model
crab_spec.parameters.covariance = results_joint.parameters.get_subcovariance(
crab_spec.parameters
)
print(crab_spec)
LogParabolaSpectralModel
name value error unit min max frozen
--------- --------- --------- -------------- --- --- ------
amplitude 3.721e-03 6.347e-04 cm-2 s-1 TeV-1 nan nan False
reference 5.055e-05 0.000e+00 TeV nan nan True
alpha 1.246e+00 3.935e-02 nan nan False
beta 6.247e-02 2.178e-03 nan nan False
We can compute flux points for Fermi-LAT and HESS datasets in order plot them together with the HAWC flux point.
[9]:
# compute Fermi-LAT and HESS flux points
e_edges = MapAxis.from_bounds(
0.01, 2.0, nbin=6, interp="log", unit="TeV"
).edges
flux_points_fermi = FluxPointsEstimator(
datasets=[dataset_fermi], e_edges=e_edges, source="Crab Nebula"
).run()
e_edges = MapAxis.from_bounds(1, 15, nbin=6, interp="log", unit="TeV").edges
flux_points_hess = FluxPointsEstimator(
datasets=[dataset_hess], e_edges=e_edges
).run()
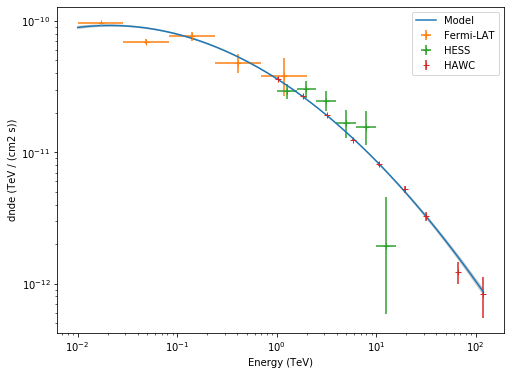
Now, Let’s plot the Crab spectrum fitted and the flux points of each instrument.
[10]:
# display spectrum and flux points
energy_range = [0.01, 120] * u.TeV
plt.figure(figsize=(8, 6))
ax = crab_spec.plot(energy_range=energy_range, energy_power=2, label="Model")
crab_spec.plot_error(ax=ax, energy_range=energy_range, energy_power=2)
flux_points_fermi.plot(ax=ax, energy_power=2, label="Fermi-LAT")
flux_points_hess.plot(ax=ax, energy_power=2, label="HESS")
flux_points_hawc.plot(ax=ax, energy_power=2, label="HAWC")
plt.legend();