This is a fixed-text formatted version of a Jupyter notebook
You can contribute with your own notebooks in this GitHub repository.
Source files: cta_data_analysis.ipynb | cta_data_analysis.py
CTA data analysis with Gammapy¶
Introduction¶
This notebook shows an example how to make a sky image and spectrum for simulated CTA data with Gammapy.
The dataset we will use is three observation runs on the Galactic center. This is a tiny (and thus quick to process and play with and learn) subset of the simulated CTA dataset that was produced for the first data challenge in August 2017.
Setup¶
As usual, we’ll start with some setup …
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
[2]:
!gammapy info --no-envvar --no-system
Gammapy package:
version : 0.15
path : /Users/adonath/github/adonath/gammapy/gammapy
Other packages:
numpy : 1.17.3
scipy : 1.3.2
astropy : 3.2.3
regions : 0.4
click : 7.0
yaml : 5.1.2
IPython : 7.9.0
jupyterlab : 1.2.3
matplotlib : 3.1.2
pandas : 0.25.3
healpy : 1.12.10
iminuit : 1.3.8
sherpa : 4.11.1
naima : 0.8.4
emcee : 3.0.2
corner : 2.0.1
parfive : 1.0.0
[3]:
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord
from astropy.convolution import Gaussian2DKernel
from regions import CircleSkyRegion
from gammapy.modeling import Fit, Datasets
from gammapy.data import DataStore
from gammapy.modeling.models import PowerLawSpectralModel, SkyModel
from gammapy.spectrum import (
SpectrumDatasetMaker,
SpectrumDataset,
FluxPointsEstimator,
FluxPointsDataset,
ReflectedRegionsBackgroundMaker,
plot_spectrum_datasets_off_regions,
)
from gammapy.maps import MapAxis, WcsNDMap, WcsGeom
from gammapy.cube import MapDatasetMaker, MapDataset, SafeMaskMaker
from gammapy.detect import TSMapEstimator, find_peaks
[4]:
# Configure the logger, so that the spectral analysis
# isn't so chatty about what it's doing.
import logging
logging.basicConfig()
log = logging.getLogger("gammapy.spectrum")
log.setLevel(logging.ERROR)
Select observations¶
A Gammapy analysis usually starts by creating a gammapy.data.DataStore and selecting observations.
This is shown in detail in the other notebook, here we just pick three observations near the galactic center.
[5]:
data_store = DataStore.from_dir("$GAMMAPY_DATA/cta-1dc/index/gps")
[6]:
# Just as a reminder: this is how to select observations
# from astropy.coordinates import SkyCoord
# table = data_store.obs_table
# pos_obs = SkyCoord(table['GLON_PNT'], table['GLAT_PNT'], frame='galactic', unit='deg')
# pos_target = SkyCoord(0, 0, frame='galactic', unit='deg')
# offset = pos_target.separation(pos_obs).deg
# mask = (1 < offset) & (offset < 2)
# table = table[mask]
# table.show_in_browser(jsviewer=True)
[7]:
obs_id = [110380, 111140, 111159]
observations = data_store.get_observations(obs_id)
[8]:
obs_cols = ["OBS_ID", "GLON_PNT", "GLAT_PNT", "LIVETIME"]
data_store.obs_table.select_obs_id(obs_id)[obs_cols]
[8]:
| OBS_ID | GLON_PNT | GLAT_PNT | LIVETIME |
|---|---|---|---|
| deg | deg | s | |
| int64 | float64 | float64 | float64 |
| 110380 | 359.9999912037958 | -1.299995937905366 | 1764.0 |
| 111140 | 358.4999833830074 | 1.3000020211954284 | 1764.0 |
| 111159 | 1.5000056568267741 | 1.299940468335294 | 1764.0 |
Make sky images¶
Define map geometry¶
Select the target position and define an ON region for the spectral analysis
[9]:
axis = MapAxis.from_edges(
np.logspace(-1.0, 1.0, 10), unit="TeV", name="energy", interp="log"
)
geom = WcsGeom.create(
skydir=(0, 0), npix=(500, 400), binsz=0.02, coordsys="GAL", axes=[axis]
)
geom
[9]:
WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (500, 400, 9)
ndim : 3
coordsys : GAL
projection : CAR
center : 0.0 deg, 0.0 deg
width : 10.0 deg x 8.0 deg
Compute images¶
Exclusion mask currently unused. Remove here or move to later in the tutorial?
[10]:
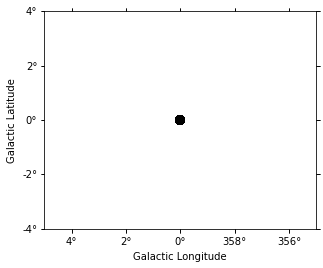
target_position = SkyCoord(0, 0, unit="deg", frame="galactic")
on_radius = 0.2 * u.deg
on_region = CircleSkyRegion(center=target_position, radius=on_radius)
[11]:
exclusion_mask = geom.to_image().region_mask([on_region], inside=False)
exclusion_mask = WcsNDMap(geom.to_image(), exclusion_mask)
exclusion_mask.plot();
[12]:
%%time
stacked = MapDataset.create(geom=geom)
maker = MapDatasetMaker(selection=["counts", "background", "exposure"])
maker_safe_mask = SafeMaskMaker(methods=["offset-max"], offset_max=2.5 * u.deg)
for obs in observations:
cutout = stacked.cutout(obs.pointing_radec, width="5 deg")
dataset = maker.run(cutout, obs)
dataset = maker_safe_mask.run(dataset, obs)
stacked.stack(dataset)
CPU times: user 2 s, sys: 402 ms, total: 2.4 s
Wall time: 2.51 s
[13]:
# The maps are cubes, with an energy axis.
# Let's also make some images:
dataset_image = stacked.to_image()
images = {
"counts": dataset_image.counts.get_image_by_idx((0,)),
"exposure": dataset_image.exposure.get_image_by_idx((0,)),
"background": dataset_image.background_model.map.get_image_by_idx((0,)),
}
images["excess"] = images["counts"] - images["background"]
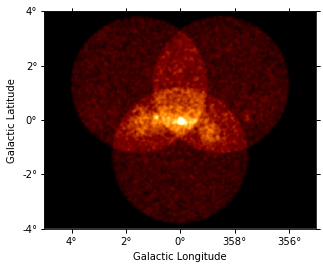
Show images¶
Let’s have a quick look at the images we computed …
[14]:
images["counts"].smooth(2).plot(vmax=5);
[15]:
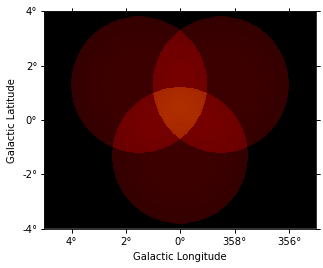
images["background"].plot(vmax=5);
[16]:
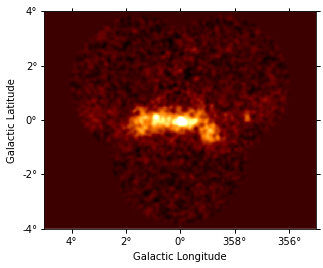
images["excess"].smooth(3).plot(vmax=2);
Source Detection¶
Use the class gammapy.detect.TSMapEstimator and gammapy.detect.find_peaks to detect sources on the images:
[17]:
kernel = Gaussian2DKernel(1, mode="oversample").array
plt.imshow(kernel);
[18]:
%%time
ts_image_estimator = TSMapEstimator()
images_ts = ts_image_estimator.run(images, kernel)
print(images_ts.keys())
dict_keys(['ts', 'sqrt_ts', 'flux', 'flux_err', 'flux_ul', 'niter'])
CPU times: user 12.7 s, sys: 270 ms, total: 13 s
Wall time: 13.5 s
[19]:
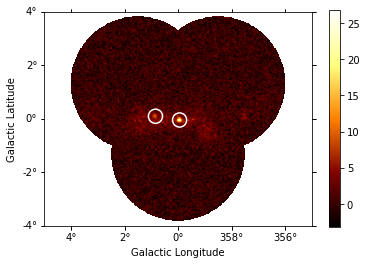
sources = find_peaks(images_ts["sqrt_ts"], threshold=8)
sources
[19]:
| value | x | y | ra | dec |
|---|---|---|---|---|
| deg | deg | |||
| float32 | int64 | int64 | float64 | float64 |
| 26.8 | 252 | 197 | 266.42400 | -29.00490 |
| 10.284 | 207 | 204 | 266.82019 | -28.16314 |
[20]:
source_pos = SkyCoord(sources["ra"], sources["dec"])
source_pos
[20]:
<SkyCoord (ICRS): (ra, dec) in deg
[(266.42399798, -29.00490483), (266.82018801, -28.16313964)]>
[21]:
# Plot sources on top of significance sky image
images_ts["sqrt_ts"].plot(add_cbar=True)
plt.gca().scatter(
source_pos.ra.deg,
source_pos.dec.deg,
transform=plt.gca().get_transform("icrs"),
color="none",
edgecolor="white",
marker="o",
s=200,
lw=1.5,
);
Spatial analysis¶
See other notebooks for how to run a 3D cube or 2D image based analysis.
Spectrum¶
We’ll run a spectral analysis using the classical reflected regions background estimation method, and using the on-off (often called WSTAT) likelihood function.
[22]:
e_reco = np.logspace(-1, np.log10(40), 40) * u.TeV
e_true = np.logspace(np.log10(0.05), 2, 200) * u.TeV
dataset_empty = SpectrumDataset.create(
e_reco=e_reco, e_true=e_true, region=on_region
)
[23]:
dataset_maker = SpectrumDatasetMaker(
containment_correction=False, selection=["counts", "aeff", "edisp"]
)
bkg_maker = ReflectedRegionsBackgroundMaker(exclusion_mask=exclusion_mask)
safe_mask_masker = SafeMaskMaker(methods=["aeff-max"], aeff_percent=10)
[24]:
%%time
datasets = []
for observation in observations:
dataset = dataset_maker.run(dataset_empty, observation)
dataset_on_off = bkg_maker.run(dataset, observation)
dataset_on_off = safe_mask_masker.run(dataset_on_off, observation)
datasets.append(dataset_on_off)
CPU times: user 5.44 s, sys: 255 ms, total: 5.69 s
Wall time: 5.99 s
[25]:
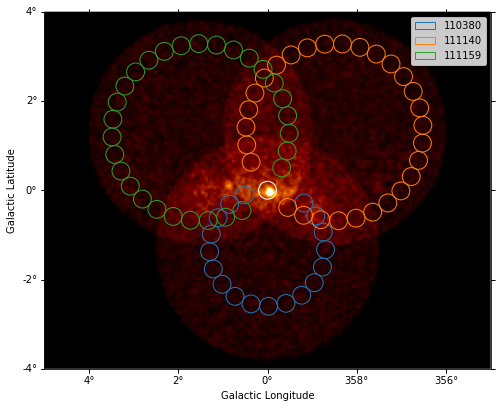
plt.figure(figsize=(8, 8))
_, ax, _ = images["counts"].smooth("0.03 deg").plot(vmax=8)
on_region.to_pixel(ax.wcs).plot(ax=ax, edgecolor="white")
plot_spectrum_datasets_off_regions(datasets, ax=ax)
Model fit¶
The next step is to fit a spectral model, using all data (i.e. a “global” fit, using all energies).
[26]:
%%time
spectral_model = PowerLawSpectralModel(
index=2, amplitude=1e-11 * u.Unit("cm-2 s-1 TeV-1"), reference=1 * u.TeV
)
model = SkyModel(spectral_model=spectral_model)
for dataset in datasets:
dataset.models = model
fit = Fit(datasets)
result = fit.run()
print(result)
OptimizeResult
backend : minuit
method : minuit
success : True
message : Optimization terminated successfully.
nfev : 108
total stat : 75.78
CPU times: user 528 ms, sys: 9.72 ms, total: 538 ms
Wall time: 597 ms
Spectral points¶
Finally, let’s compute spectral points. The method used is to first choose an energy binning, and then to do a 1-dim likelihood fit / profile to compute the flux and flux error.
[27]:
# Flux points are computed on stacked observation
stacked_dataset = Datasets(datasets).stack_reduce()
print(stacked_dataset)
SpectrumDatasetOnOff
Name : 110380
Total counts : 441
Total predicted counts : 1117.40
Total off counts : 2278.00
Total background counts : 91.22
Effective area min : 1.88e+08 cm2
Effective area max : 4.64e+10 cm2
Livetime : 5.29e+03 s
Number of total bins : 39
Number of fit bins : 29
Fit statistic type : wstat
Fit statistic value (-2 log(L)) : 29.84
Number of parameters : 3
Number of free parameters : 2
Model type : SkyModels
Acceptance mean: : 1.0
[28]:
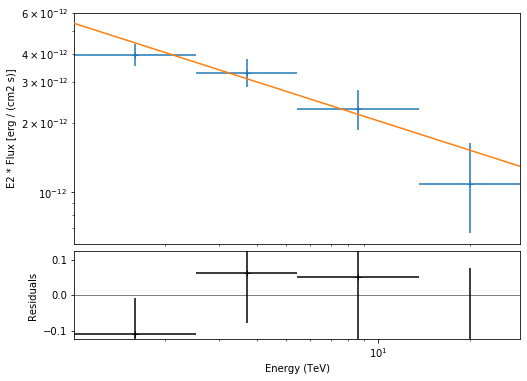
e_edges = np.logspace(0, 1.5, 5) * u.TeV
stacked_dataset.model = model
fpe = FluxPointsEstimator(datasets=[stacked_dataset], e_edges=e_edges)
flux_points = fpe.run()
flux_points.table_formatted
[28]:
| e_ref | e_min | e_max | ref_dnde | ref_flux | ref_eflux | ref_e2dnde | norm | stat | norm_err | counts [1] | norm_errp | norm_errn | norm_ul | sqrt_ts | ts | norm_scan [11] | stat_scan [11] | dnde | dnde_ul | dnde_err | dnde_errp | dnde_errn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TeV | TeV | TeV | 1 / (cm2 s TeV) | 1 / (cm2 s) | TeV / (cm2 s) | TeV / (cm2 s) | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) | |||||||||||
| float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1.588 | 1.002 | 2.518 | 1.101e-12 | 1.730e-12 | 2.576e-12 | 2.777e-12 | 0.890 | 5.783 | 0.099 | 124 | 0.103 | 0.095 | 1.101 | 13.997 | 195.910 | 0.200 .. 5.000 | 92.974 .. 547.190 | 9.798e-13 | 1.212e-12 | 1.089e-13 | 1.130e-13 | 1.050e-13 |
| 3.697 | 2.518 | 5.429 | 1.422e-13 | 4.242e-13 | 1.500e-12 | 1.944e-12 | 1.061 | 1.271 | 0.145 | 72 | 0.151 | 0.139 | 1.374 | 12.302 | 151.342 | 0.200 .. 5.000 | 72.914 .. 256.695 | 1.509e-13 | 1.954e-13 | 2.058e-14 | 2.145e-14 | 1.973e-14 |
| 8.607 | 5.429 | 13.647 | 1.836e-14 | 1.563e-13 | 1.262e-12 | 1.360e-12 | 1.052 | 12.004 | 0.206 | 35 | 0.218 | 0.195 | 1.512 | 8.483 | 71.954 | 0.200 .. 5.000 | 46.142 .. 137.818 | 1.931e-14 | 2.777e-14 | 3.788e-15 | 4.008e-15 | 3.575e-15 |
| 20.037 | 13.647 | 29.419 | 2.371e-15 | 3.834e-14 | 7.345e-13 | 9.520e-13 | 0.717 | 3.603 | 0.318 | 7 | 0.358 | 0.280 | 1.520 | 3.733 | 13.937 | 0.200 .. 5.000 | 8.306 .. 49.762 | 1.700e-15 | 3.605e-15 | 7.533e-16 | 8.494e-16 | 6.628e-16 |
Plot¶
Let’s plot the spectral model and points. You could do it directly, but there is a helper class. Note that a spectral uncertainty band, a “butterfly” is drawn, but it is very thin, i.e. barely visible.
[29]:
model.spectral_model.parameters.covariance = result.parameters.covariance
flux_points_dataset = FluxPointsDataset(data=flux_points, models=model)
[30]:
plt.figure(figsize=(8, 6))
flux_points_dataset.peek();
Exercises¶
Re-run the analysis above, varying some analysis parameters, e.g.
Select a few other observations
Change the energy band for the map
Change the spectral model for the fit
Change the energy binning for the spectral points
Change the target. Make a sky image and spectrum for your favourite source.
If you don’t know any, the Crab nebula is the “hello world!” analysis of gamma-ray astronomy.
[31]:
# print('hello world')
# SkyCoord.from_name('crab')
What next?¶
This notebook showed an example of a first CTA analysis with Gammapy, using simulated 1DC data.
Let us know if you have any question or issues!