Note
Go to the end to download the full example code or to run this example in your browser via Binder
Flux Profile Estimation#
Learn how to estimate flux profiles on a Fermi-LAT dataset.
Prerequisites#
Knowledge of 3D data reduction and datasets used in Gammapy, see for instance the first analysis tutorial.
Context#
A useful tool to study and compare the saptial distribution of flux in images and data cubes is the measurement of flxu profiles. Flux profiles can show spatial correlations of gamma-ray data with e.g. gas maps or other type of gamma-ray data. Most commonly flux profiles are measured along some preferred coordinate axis, either radially distance from a source of interest, along longitude and latitude coordinate axes or along the path defined by two spatial coordinates.
Proposed Approach#
Flux profile estimation essentially works by estimating flux points for a set of predefined spatially connected regions. For radial flux profiles the shape of the regions are annuli with a common center, for linear profiles it’s typically a rectangular shape.
We will work on a pre-computed MapDataset of Fermi-LAT data, use
SkyRegion to define the structure of the bins of the flux profile and
run the actually profile extraction using the FluxProfileEstimator
import numpy as np
from astropy import units as u
from astropy.coordinates import SkyCoord
# %matplotlib inline
import matplotlib.pyplot as plt
Setup#
from IPython.display import display
from gammapy.datasets import MapDataset
from gammapy.estimators import FluxPoints, FluxProfileEstimator
from gammapy.maps import RegionGeom
from gammapy.modeling.models import PowerLawSpectralModel
Check setup#
from gammapy.utils.check import check_tutorials_setup
from gammapy.utils.regions import (
make_concentric_annulus_sky_regions,
make_orthogonal_rectangle_sky_regions,
)
check_tutorials_setup()
System:
python_executable : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/bin/python
python_version : 3.9.16
machine : x86_64
system : Linux
Gammapy package:
version : 1.0.1
path : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/gammapy
Other packages:
numpy : 1.24.2
scipy : 1.10.1
astropy : 5.2.1
regions : 0.7
click : 8.1.3
yaml : 6.0
IPython : 8.11.0
jupyterlab : not installed
matplotlib : 3.7.1
pandas : not installed
healpy : 1.16.2
iminuit : 2.21.0
sherpa : 4.15.0
naima : 0.10.0
emcee : 3.1.4
corner : 2.2.1
Gammapy environment variables:
GAMMAPY_DATA : /home/runner/work/gammapy-docs/gammapy-docs/gammapy-datasets/1.0.1
Read and Introduce Data#
dataset = MapDataset.read(
"$GAMMAPY_DATA/fermi-3fhl-gc/fermi-3fhl-gc.fits.gz", name="fermi-dataset"
)
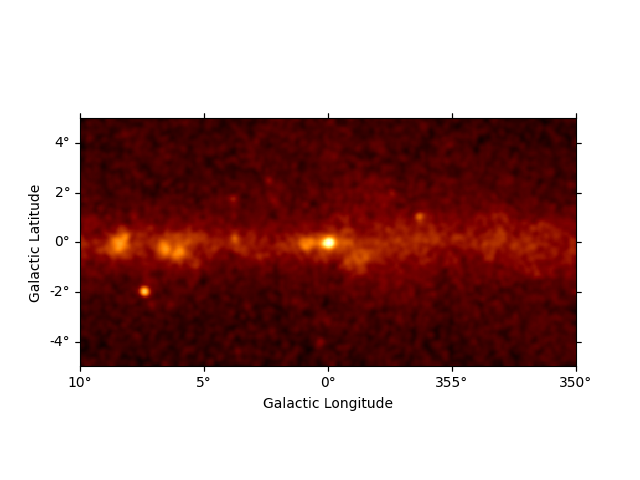
This is what the counts image we will work with looks like:
counts_image = dataset.counts.sum_over_axes()
counts_image.smooth("0.1 deg").plot(stretch="sqrt")
<WCSAxes: >
There are 400x200 pixels in the dataset and 11 energy bins between 10 GeV and 2 TeV:
print(dataset.counts)
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (400, 200, 11)
ndim : 3
unit :
dtype : >i4
Profile Estimation#
Configuration#
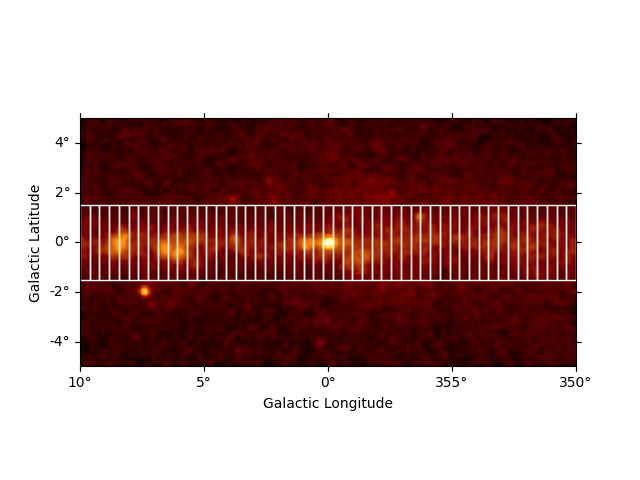
We start by defining a list of spatially connected regions along the
galactic longitude axis. For this there is a helper function
make_orthogonal_rectangle_sky_regions. The individual region bins
for the profile have a height of 3 deg and in total there are 31 bins.
The starts from lon = 10 deg tand goes to lon = 350 deg. In addition we
have to specify the wcs to take into account possible projections
effects on the region definition:
regions = make_orthogonal_rectangle_sky_regions(
start_pos=SkyCoord("10d", "0d", frame="galactic"),
end_pos=SkyCoord("350d", "0d", frame="galactic"),
wcs=counts_image.geom.wcs,
height="3 deg",
nbin=51,
)
We can use the RegionGeom object to illustrate the regions on top of
the counts image:
plt.figure()
geom = RegionGeom.create(region=regions)
ax = counts_image.smooth("0.1 deg").plot(stretch="sqrt")
geom.plot_region(ax=ax, color="w")
/home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/regions/shapes/rectangle.py:208: UserWarning: Setting the 'color' property will override the edgecolor or facecolor properties.
return Rectangle(xy=xy, width=width, height=height,
<WCSAxes: >
Next we create the FluxProfileEstimator. For the estimation of the
flux profile we assume a spectral model with a power-law shape and an
index of 2.3
flux_profile_estimator = FluxProfileEstimator(
regions=regions,
spectrum=PowerLawSpectralModel(index=2.3),
energy_edges=[10, 2000] * u.GeV,
selection_optional=["ul"],
)
We can see the full configuration by printing the estimator object:
print(flux_profile_estimator)
FluxProfileEstimator
--------------------
energy_edges : [ 10. 2000.] GeV
fit : <gammapy.modeling.fit.Fit object at 0x7f31817ccca0>
n_sigma : 1
n_sigma_ul : 2
norm_max : 5
norm_min : 0.2
norm_n_values : 11
norm_values : None
null_value : 0
reoptimize : False
selection_optional : ['ul']
source : 0
spectrum : PowerLawSpectralModel
sum_over_energy_groups : False
Run Estimation#
Now we can run the profile estimation and explore the results:
profile = flux_profile_estimator.run(datasets=dataset)
print(profile)
FluxPoints
----------
geom : RegionGeom
axes : ['lon', 'lat', 'energy', 'projected-distance']
shape : (1, 1, 1, 51)
quantities : ['norm', 'norm_err', 'norm_ul', 'ts', 'npred', 'npred_excess', 'stat', 'counts', 'success']
ref. model : pl
n_sigma : 1
n_sigma_ul : 2
sqrt_ts_threshold_ul : 2
sed type init : likelihood
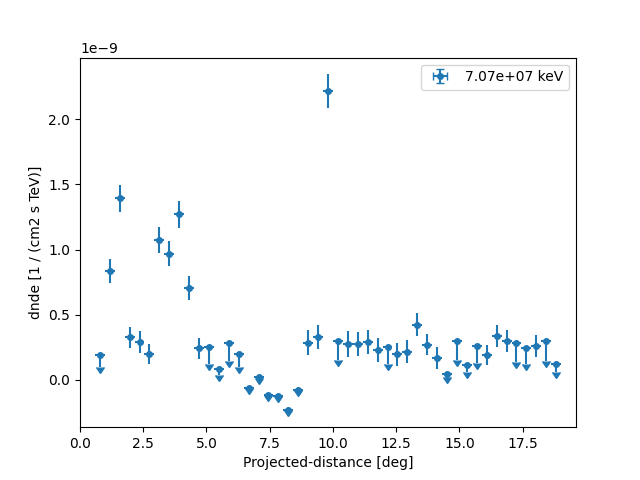
We can see the flux profile is represented by a FluxPoints object
with a projected-distance axis, which defines the main axis the flux
profile is measured along. The lon and lat axes can be ignored.
Plotting Results#
Let us directly plot the result using plot:
plt.figure()
ax = profile.plot(sed_type="dnde")
ax.set_yscale("linear")
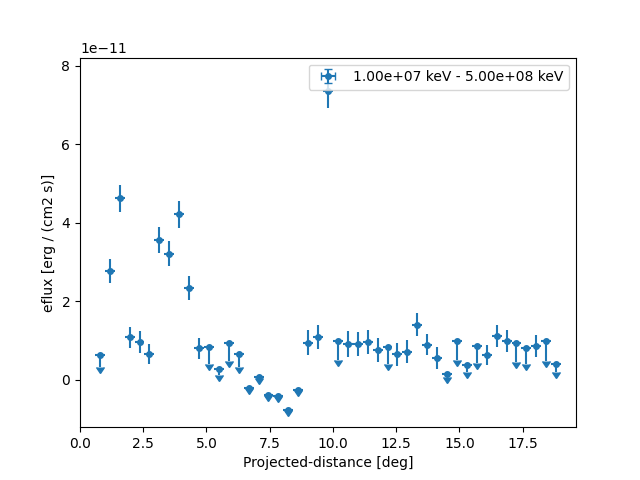
Based on the spectral model we specified above we can also plot in any other sed type, e.g. energy flux and define a different threshold when to plot upper limits:
profile.sqrt_ts_threshold_ul = 2
plt.figure()
ax = profile.plot(sed_type="eflux")
ax.set_yscale("linear")
We can also plot any other quantity of interest, that is defined on the
FluxPoints result object. E.g. the predicted total counts,
background counts and excess counts:
quantities = ["npred", "npred_excess", "npred_background"]
fig, ax = plt.subplots()
for quantity in quantities:
profile[quantity].plot(ax=ax, label=quantity.title())
ax.set_ylabel("Counts")
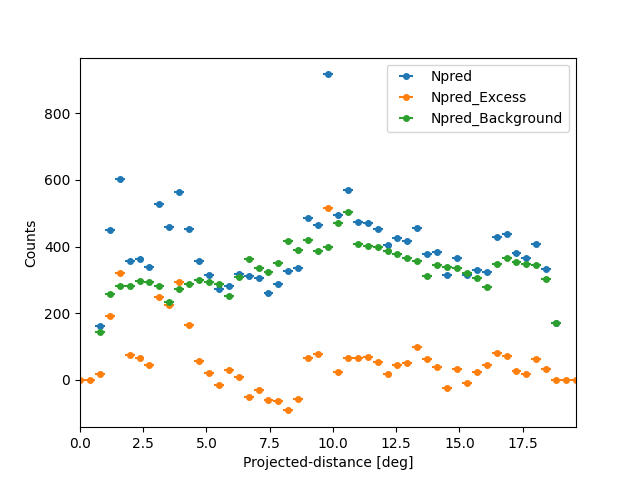
Text(38.347222222222214, 0.5, 'Counts')
Serialisation and I/O#
The profile can be serialised using write, given a
specific format:
profile.write(
filename="flux_profile_fermi.fits",
format="profile",
overwrite=True,
sed_type="dnde",
)
profile_new = FluxPoints.read(filename="flux_profile_fermi.fits", format="profile")
fig = plt.figure()
ax = profile_new.plot()
ax.set_yscale("linear")
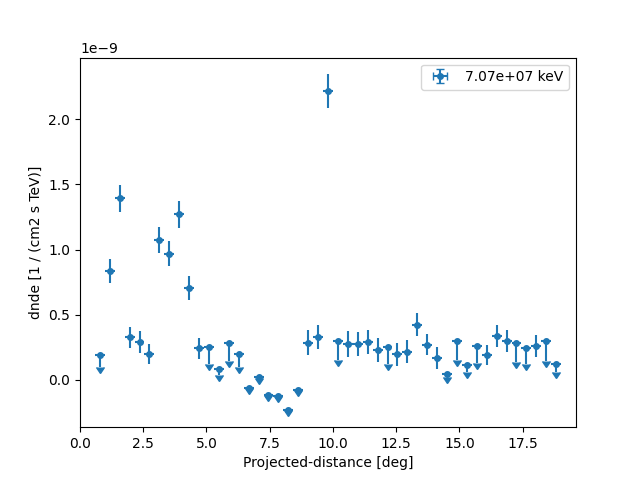
The profile can be serialised to a Table object
using:
table = profile.to_table(format="profile", formatted=True)
display(table)
x_min x_max x_ref ... counts success
deg deg deg ...
-------------------- ------------------- ------------------ ... ------ -------
-0.19607843137254918 0.19607843137254918 0.0 ... 0.0 False
0.19607843137254918 0.5882352941176466 0.3921568627450979 ... 0.0 False
0.5882352941176466 0.9803921568627441 0.7843137254901953 ... 163.0 True
0.9803921568627441 1.3725490196078436 1.176470588235294 ... 448.0 True
1.3725490196078436 1.7647058823529418 1.5686274509803928 ... 599.0 True
1.7647058823529418 2.1568627450980395 1.9607843137254908 ... 354.0 True
2.1568627450980395 2.549019607843138 2.3529411764705888 ... 367.0 True
2.549019607843138 2.941176470588236 2.745098039215687 ... 339.0 True
2.941176470588236 3.3333333333333344 3.137254901960785 ... 531.0 True
3.3333333333333344 3.725490196078432 3.529411764705883 ... 458.0 True
... ... ... ... ... ...
15.882352941176464 16.274509803921596 16.07843137254903 ... 321.0 True
16.274509803921596 16.666666666666696 16.470588235294144 ... 421.0 True
16.666666666666696 17.058823529411775 16.862745098039234 ... 431.0 True
17.058823529411775 17.4509803921569 17.254901960784338 ... 374.0 True
17.4509803921569 17.843137254902004 17.647058823529452 ... 370.0 True
17.843137254902004 18.23529411764708 18.03921568627454 ... 410.0 True
18.23529411764708 18.627450980392158 18.43137254901962 ... 336.0 True
18.627450980392158 19.01960784313726 18.82352941176471 ... 172.0 True
19.01960784313726 19.41176470588239 19.215686274509824 ... 0.0 False
19.41176470588239 19.8039215686275 19.607843137254946 ... 0.0 False
Length = 51 rows
No we can also estimate a radial profile starting from the Galactic center:
regions = make_concentric_annulus_sky_regions(
center=SkyCoord("0d", "0d", frame="galactic"),
radius_max="1.5 deg",
nbin=11,
)
Again we first illustrate the regions:
plt.figure()
geom = RegionGeom.create(region=regions)
gc_image = counts_image.cutout(
position=SkyCoord("0d", "0d", frame="galactic"), width=3 * u.deg
)
ax = gc_image.smooth("0.1 deg").plot(stretch="sqrt")
geom.plot_region(ax=ax, color="w")
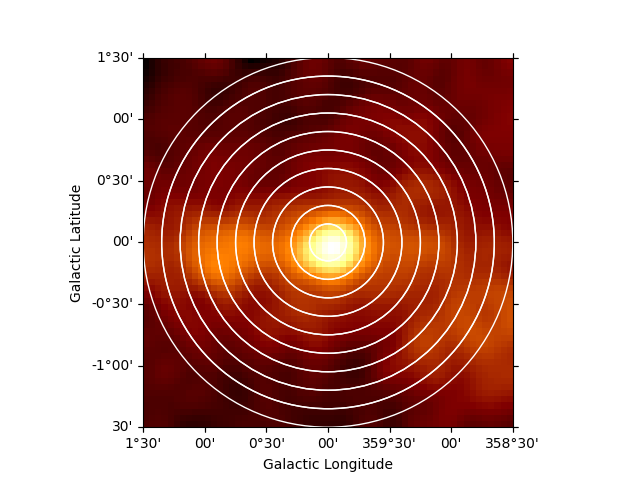
/home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/regions/core/compound.py:160: UserWarning: Setting the 'color' property will override the edgecolor or facecolor properties.
patch = mpatches.PathPatch(path, **mpl_kwargs)
<WCSAxes: >
This time we define two energy bins and include the fit statistic profile in the computation:
flux_profile_estimator = FluxProfileEstimator(
regions=regions,
spectrum=PowerLawSpectralModel(index=2.3),
energy_edges=[10, 100, 2000] * u.GeV,
selection_optional=["ul", "scan"],
norm_values=np.linspace(-1, 5, 11),
)
profile = flux_profile_estimator.run(datasets=dataset)
We can directly plot the result:
plt.figure()
profile.plot(axis_name="projected-distance", sed_type="flux")
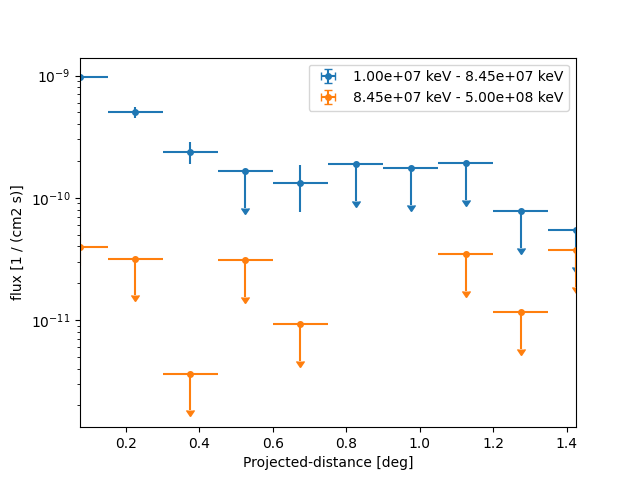
<Axes: xlabel='Projected-distance [deg]', ylabel='flux [1 / (cm2 s)]'>
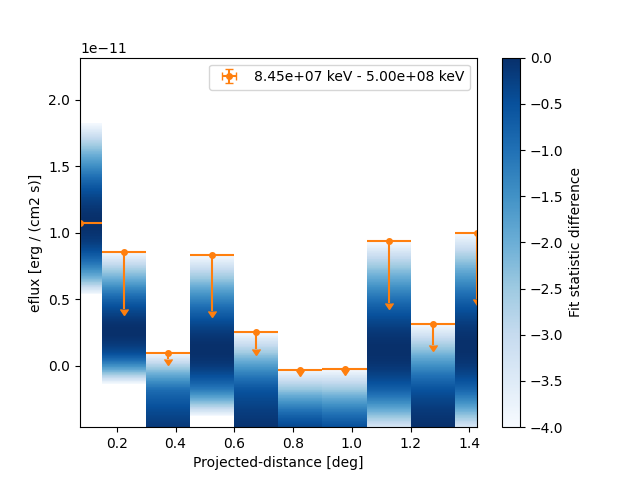
However because of the powerlaw spectrum the flux at high energies is much lower. To extract the profile at high energies only we can use:
profile_high = profile.slice_by_idx({"energy": slice(1, 2)})
And now plot the points together with the likelihood profiles:
fig, ax = plt.subplots()
profile_high.plot(ax=ax, sed_type="eflux", color="tab:orange")
profile_high.plot_ts_profiles(ax=ax, sed_type="eflux")
ax.set_yscale("linear")
plt.show()
# sphinx_gallery_thumbnail_number = 2
Total running time of the script: ( 0 minutes 25.724 seconds)