This is a fixed-text formatted version of a Jupyter notebook
- Try online
- You can contribute with your own notebooks in this GitHub repository.
- Source files: hess.ipynb | hess.py
Analysis of H.E.S.S. DL3 data with Gammapy¶
In September 2018 the H.E.S.S. collaboration released a small subset of archival data in FITS format. This tutorial explains how to analyse this data with Gammapy. We will analyse four observation runs of the Crab nebula, which are part of the H.E.S.S. first public test data release. The data was release without corresponding background models. In background_model.ipynb we show how to make a simple background model, which is also used in this tutorial. The background model is not perfect; it assumes radial symmetry and is in general derived from only a few observations, but still good enough for a reliable analysis > 1TeV.
Note: The high level Analysis class is a new feature added in Gammapy v0.14. In the curret state it supports the standard analysis cases of a joint or stacked 3D and 1D analysis. It provides only limited access to analaysis parameters via the config file. It is expected that the format of the YAML config will be extended and change in future Gammapy versions.
We will first show how to configure and run a stacked 3D analysis and then address the classical spectral analysis using reflected regions later. The structure of the tutorial follows a typical analysis:
- Analysis configuration
- Observation slection
- Data reduction
- Model fitting
- Estimating flux points
Finally we will compare the results against a reference model.
Setup¶
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
[2]:
import yaml
from pathlib import Path
from regions import CircleSkyRegion
from astropy import units as u
from astropy.coordinates import SkyCoord
from gammapy.scripts import Analysis, AnalysisConfig
from gammapy.modeling.models import create_crab_spectral_model
Analysis configuration¶
For configuration of the analysis we use the YAML data format. YAML is a machine readable serialisation format, that is also friendly for humans to read. In this tutorial we will write the configuration file just using Python strings, but of course the file can be created and modified with any text editor of your choice.
Here is what the configuration for our analysis looks like:
[3]:
config_str = """
general:
logging:
level: INFO
outdir: .
observations:
datastore: $GAMMAPY_DATA/hess-dl3-dr1/hess-dl3-dr3-with-background.fits.gz
filters:
- filter_type: par_value
value_param: Crab
variable: TARGET_NAME
datasets:
dataset-type: MapDataset
stack-datasets: true
offset-max: 2.5 deg
geom:
skydir: [83.633, 22.014]
width: [5, 5]
binsz: 0.02
coordsys: CEL
proj: TAN
axes:
- name: energy
hi_bnd: 10
lo_bnd: 1
nbin: 5
interp: log
node_type: edges
unit: TeV
fit:
fit_range:
max: 30 TeV
min: 1 TeV
flux-points:
fp_binning:
lo_bnd: 1
hi_bnd: 10
interp: log
nbin: 3
unit: TeV
"""
We first create an AnalysiConfig object from it:
[4]:
config = AnalysisConfig(config_str)
Observation selection¶
Now we create the high level Analysis object from the config object:
[5]:
analysis = Analysis(config)
INFO:gammapy.scripts.analysis:Setting logging config: {'level': 'INFO'}
And directly select and load the observatiosn from disk using .get_observations():
[6]:
analysis.get_observations()
INFO:gammapy.scripts.analysis:Fetching observations.
INFO:gammapy.scripts.analysis:Info for OBS_ID = 23592
- Start time: 53347.91
- Pointing pos: RA 82.01 deg / Dec 22.01 deg
- Observation duration: 1686.0 s
- Dead-time fraction: 6.212 %
INFO:gammapy.scripts.analysis:Info for OBS_ID = 23523
- Start time: 53343.92
- Pointing pos: RA 83.63 deg / Dec 21.51 deg
- Observation duration: 1687.0 s
- Dead-time fraction: 6.240 %
INFO:gammapy.scripts.analysis:Info for OBS_ID = 23526
- Start time: 53343.95
- Pointing pos: RA 83.63 deg / Dec 22.51 deg
- Observation duration: 1683.0 s
- Dead-time fraction: 6.555 %
INFO:gammapy.scripts.analysis:Info for OBS_ID = 23559
- Start time: 53345.96
- Pointing pos: RA 85.25 deg / Dec 22.01 deg
- Observation duration: 1686.0 s
- Dead-time fraction: 6.398 %
The observations are now availabe on the Analysis object. The selection corresponds to the following ids:
[7]:
analysis.observations.ids
[7]:
['23592', '23523', '23526', '23559']
Now we can access and inspect individual observations by accessing with the observation id:
[8]:
print(analysis.observations["23592"])
Info for OBS_ID = 23592
- Start time: 53347.91
- Pointing pos: RA 82.01 deg / Dec 22.01 deg
- Observation duration: 1686.0 s
- Dead-time fraction: 6.212 %
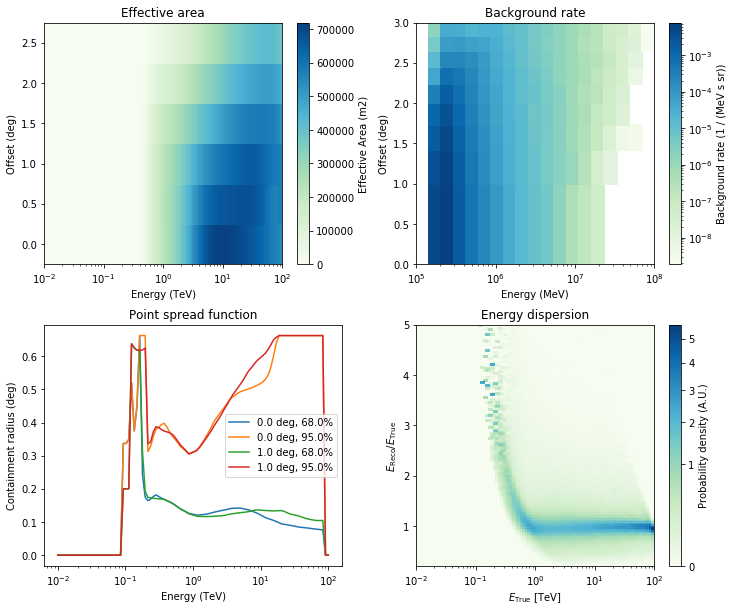
And also show a few overview plots using the .peek() method:
[9]:
analysis.observations["23592"].peek()
WARNING:gammapy.irf.psf_table:PSF does not integrate to unity within a precision of 1% in each energy bin. Containment radius computation might give biased results.
WARNING:gammapy.irf.psf_table:PSF does not integrate to unity within a precision of 1% in each energy bin. Containment radius computation might give biased results.
WARNING:gammapy.irf.psf_table:PSF does not integrate to unity within a precision of 1% in each energy bin. Containment radius computation might give biased results.
WARNING:gammapy.irf.psf_table:PSF does not integrate to unity within a precision of 1% in each energy bin. Containment radius computation might give biased results.
Data reduction¶
Now we proceed to the data reduction. In the config file we have chosen a WCS map geometry, energy axis and decided to stack the maps. We can run the reduction using .get_datasets():
[10]:
%%time
analysis.get_datasets()
INFO:gammapy.scripts.analysis:Creating geometry.
INFO:gammapy.scripts.analysis:Creating datasets.
CPU times: user 6.8 s, sys: 609 ms, total: 7.41 s
Wall time: 7.55 s
As we have chosen to stack the data, there is finally one dataset contained:
[11]:
analysis.datasets.names
[11]:
['stacked']
We can print the dataset as well:
[12]:
print(analysis.datasets["stacked"])
MapDataset
Name : stacked
Total counts : 7474
Total predicted counts : 2326.93
Total background counts : 2326.93
Exposure min : 5.97e+07 m2 s
Exposure max : 3.15e+09 m2 s
Number of total bins : 312500
Number of fit bins : 309230
Fit statistic type : cash
Fit statistic value (-2 log(L)) : 73346.62
Number of models : 0
Number of parameters : 3
Number of free parameters : 1
Component 0:
Name : background
Type : BackgroundModel
Parameters:
norm : 1.000
tilt (frozen) : 0.000
reference (frozen) : 1.000 TeV
As you can see the dataset comes with a predefined background model out of the data reduction, but no source model has been set yet.
The counts, exposure and background model maps are directly available on the dataset and can be printed and plotted:
[13]:
counts = analysis.datasets["stacked"].counts
[14]:
print(counts)
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (250, 250, 5)
ndim : 3
unit :
dtype : float32
[15]:
counts.smooth("0.05 deg").plot_interactive()

Model fitting¶
Now we define a model to be fitted to the dataset:
[16]:
model_config = """
components:
- name: crab
type: SkyModel
spatial:
type: PointSpatialModel
frame: icrs
parameters:
- name: lon_0
value: 83.63
unit: deg
- name: lat_0
value: 22.14
unit: deg
spectral:
type: PowerLawSpectralModel
parameters:
- name: amplitude
value: 1.0e-12
unit: cm-2 s-1 TeV-1
- name: index
value: 2.0
unit: ''
- name: reference
value: 1.0
unit: TeV
frozen: true
"""
Now we set the model on the analysis object:
[17]:
analysis.set_model(model_config)
INFO:gammapy.scripts.analysis:Reading model.
INFO:gammapy.scripts.analysis:SkyModels
Component 0: SkyModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
lon_0 8.363e+01 nan deg nan nan False
lat_0 2.214e+01 nan deg nan nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 nan nan False
index 2.000e+00 nan nan nan False
reference 1.000e+00 nan TeV nan nan True
[18]:
print(analysis.model)
SkyModels
Component 0: SkyModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
lon_0 8.363e+01 nan deg nan nan False
lat_0 2.214e+01 nan deg nan nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 nan nan False
index 2.000e+00 nan nan nan False
reference 1.000e+00 nan TeV nan nan True
[19]:
print(analysis.model["crab"])
SkyModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
lon_0 8.363e+01 nan deg nan nan False
lat_0 2.214e+01 nan deg nan nan False
amplitude 1.000e-12 nan cm-2 s-1 TeV-1 nan nan False
index 2.000e+00 nan nan nan False
reference 1.000e+00 nan TeV nan nan True
Finally we run the fit:
[20]:
analysis.run_fit()
INFO:gammapy.scripts.analysis:Fitting reduced datasets.
INFO:gammapy.scripts.analysis:OptimizeResult
backend : minuit
method : minuit
success : True
message : Optimization terminated successfully.
nfev : 312
total stat : 64490.20
[21]:
print(analysis.fit_result)
OptimizeResult
backend : minuit
method : minuit
success : True
message : Optimization terminated successfully.
nfev : 312
total stat : 64490.20
This is how we can write the model back to file again:
[22]:
analysis.model.to_yaml("model-best-fit.yaml")
[23]:
!cat model-best-fit.yaml
components:
- name: crab
type: SkyModel
spatial:
type: PointSpatialModel
frame: icrs
parameters:
- name: lon_0
value: 83.61894929306811
unit: deg
- name: lat_0
value: 22.024836258944713
unit: deg
spectral:
type: PowerLawSpectralModel
parameters:
- name: amplitude
value: 6.290310266334393e-11
unit: cm-2 s-1 TeV-1
- name: index
value: 2.785083946374748
unit: ''
- name: reference
value: 1.0
unit: TeV
frozen: true
Inspecting residuals¶
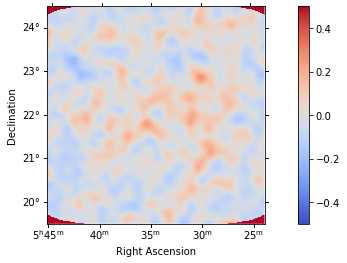
For any fit it is usefull to inspect the residual images. We have a few option on the dataset object to handle this. First we can use .plot_residuals() to plot a residual image, summed over all energies:
[24]:
analysis.datasets["stacked"].plot_residuals(
method="diff/sqrt(model)", vmin=-0.5, vmax=0.5
);
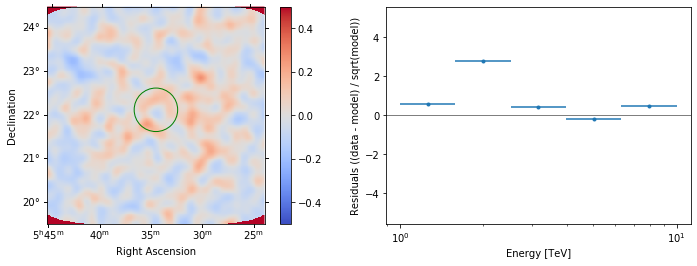
In addition we can aslo specify a region in the map to show the spectral residuals:
[25]:
region = CircleSkyRegion(
center=SkyCoord("83.63 deg", "22.14 deg"), radius=0.5 * u.deg
)
[26]:
analysis.datasets["stacked"].plot_residuals(
region=region, method="diff/sqrt(model)", vmin=-0.5, vmax=0.5
);
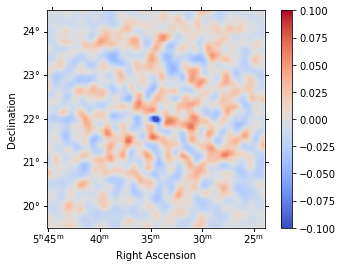
We can also directly access the .residuals() to get a map, that we can plot interactively:
[27]:
residuals = analysis.datasets["stacked"].residuals(method="diff")
residuals.smooth("0.08 deg").plot_interactive(
cmap="coolwarm", vmin=-0.1, vmax=0.1, stretch="linear", add_cbar=True
)
Inspecting likelihood profiles¶
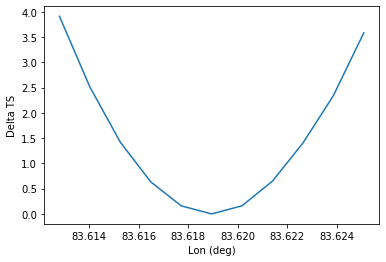
To check the quality of the fit it is also useful to plot likelihood profiles for specific parameters. For this we use analysis.fit.likelihood_profile()
[28]:
profile = analysis.fit.likelihood_profile(parameter="lon_0")
For a good fit and error estimate the profile should be parabolic, if we plot it:
[29]:
total_stat = analysis.fit_result.total_stat
plt.plot(profile["values"], profile["likelihood"] - total_stat)
plt.xlabel("Lon (deg)")
plt.ylabel("Delta TS")
[29]:
Text(0, 0.5, 'Delta TS')
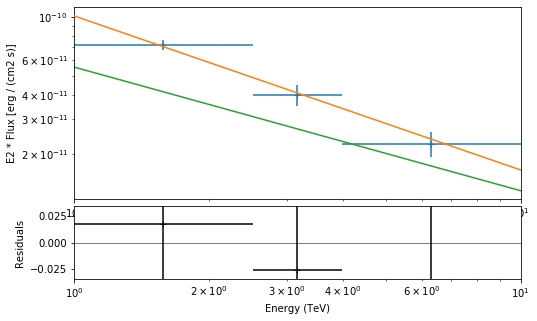
Flux points¶
[30]:
analysis.get_flux_points(source="crab")
INFO:gammapy.scripts.analysis:Calculating flux points.
INFO:gammapy.scripts.analysis:
e_ref ref_flux ... dnde_err is_ul
TeV 1 / (cm2 s) ... 1 / (cm2 s TeV)
------------------ ---------------------- ... ---------------------- -----
1.5848931924611136 2.8430798565965568e-11 ... 1.0462744900578112e-12 False
3.1622776601683795 3.815367550602265e-12 ... 3.028947216617177e-13 False
6.309573444801933 2.4140079194837277e-12 ... 4.951361673757777e-14 False
[31]:
plt.figure(figsize=(8, 5))
ax_sed, ax_residuals = analysis.flux_points.peek()
crab_spectrum = create_crab_spectral_model("hess_pl")
crab_spectrum.plot(
ax=ax_sed,
energy_range=[1, 10] * u.TeV,
energy_power=2,
flux_unit="erg-1 cm-2 s-1",
)
[31]:
<matplotlib.axes._subplots.AxesSubplot at 0x1289576d8>
Exercises¶
- Run a spectral analysis using reflected regions without stacking the datasets. You can use
AnalysisConfig.from_template("1d")to get an example configuration file. Add the resulting flux points to the SED plotted above.