This is a fixed-text formatted version of a Jupyter notebook
- Try online
- You can contribute with your own notebooks in this GitHub repository.
- Source files: sed_fitting_gammacat_fermi.ipynb | sed_fitting_gammacat_fermi.py
Flux point fitting in Gammapy¶
Introduction¶
In this tutorial we’re going to learn how to fit spectral models to combined Fermi-LAT and IACT flux points.
The central class we’re going to use for this example analysis is:
In addition we will work with the following data classes:
- gammapy.spectrum.FluxPoints
- gammapy.catalog.SourceCatalogGammaCat
- gammapy.catalog.SourceCatalog3FHL
- gammapy.catalog.SourceCatalog3FGL
And the following spectral model classes:
Setup¶
Let us start with the usual IPython notebook and Python imports:
[1]:
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
[2]:
from astropy import units as u
from gammapy.modeling.models import (
PowerLawSpectralModel,
ExpCutoffPowerLawSpectralModel,
LogParabolaSpectralModel,
)
from gammapy.spectrum import FluxPointsDataset, FluxPoints
from gammapy.catalog import (
SourceCatalog3FGL,
SourceCatalogGammaCat,
SourceCatalog3FHL,
)
from gammapy.modeling import Fit
Load spectral points¶
For this analysis we choose to work with the source ‘HESS J1507-622’ and the associated Fermi-LAT sources ‘3FGL J1506.6-6219’ and ‘3FHL J1507.9-6228e’. We load the source catalogs, and then access source of interest by name:
[3]:
fermi_3fgl = SourceCatalog3FGL()
fermi_3fhl = SourceCatalog3FHL()
gammacat = SourceCatalogGammaCat("$GAMMAPY_DATA/gamma-cat/gammacat.fits.gz")
[4]:
source_gammacat = gammacat["HESS J1507-622"]
source_fermi_3fgl = fermi_3fgl["3FGL J1506.6-6219"]
source_fermi_3fhl = fermi_3fhl["3FHL J1507.9-6228e"]
The corresponding flux points data can be accessed with .flux_points attribute:
[5]:
flux_points_gammacat = source_gammacat.flux_points
flux_points_gammacat.table
[5]:
| e_ref | dnde | dnde_errn | dnde_errp |
|---|---|---|---|
| TeV | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) | 1 / (cm2 s TeV) |
| float32 | float32 | float32 | float32 |
| 0.8609004 | 2.29119e-12 | 8.705427e-13 | 8.955021e-13 |
| 1.561512 | 6.981717e-13 | 2.203541e-13 | 2.304066e-13 |
| 2.763753 | 1.690615e-13 | 6.758698e-14 | 7.188384e-14 |
| 4.891597 | 7.729249e-14 | 2.401318e-14 | 2.607487e-14 |
| 9.988584 | 1.032534e-14 | 5.063147e-15 | 5.641954e-15 |
| 27.04035 | 7.449867e-16 | 5.72089e-16 | 7.259987e-16 |
In the Fermi-LAT catalogs, integral flux points are given. Currently the flux point fitter only works with differential flux points, so we apply the conversion here.
[6]:
flux_points_3fgl = source_fermi_3fgl.flux_points.to_sed_type(
sed_type="dnde", model=source_fermi_3fgl.spectral_model
)
flux_points_3fhl = source_fermi_3fhl.flux_points.to_sed_type(
sed_type="dnde", model=source_fermi_3fhl.spectral_model
)
Finally we stack the flux points into a single FluxPoints object and drop the upper limit values, because currently we can’t handle them in the fit:
[7]:
# stack flux point tables
flux_points = FluxPoints.stack(
[flux_points_gammacat, flux_points_3fhl, flux_points_3fgl]
)
# drop the flux upper limit values
flux_points = flux_points.drop_ul()
Power Law Fit¶
First we start with fitting a simple power law.
[8]:
pwl = PowerLawSpectralModel(
index=2, amplitude="1e-12 cm-2 s-1 TeV-1", reference="1 TeV"
)
After creating the model we run the fit by passing the 'flux_points' and 'pwl' objects:
[9]:
dataset_pwl = FluxPointsDataset(pwl, flux_points, likelihood="chi2assym")
fitter = Fit(dataset_pwl)
result_pwl = fitter.run()
And print the result:
[10]:
print(result_pwl)
OptimizeResult
backend : minuit
method : minuit
success : True
message : Optimization terminated successfully.
nfev : 40
total stat : 33.68
[11]:
print(pwl)
PowerLawSpectralModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
index 1.966e+00 nan nan nan False
amplitude 1.345e-12 nan cm-2 s-1 TeV-1 nan nan False
reference 1.000e+00 nan TeV nan nan True
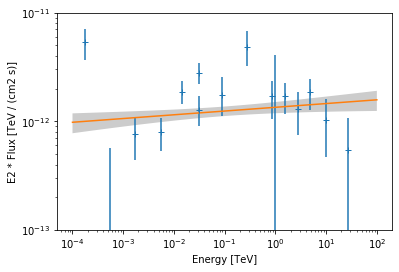
Finally we plot the data points and the best fit model:
[12]:
ax = flux_points.plot(energy_power=2)
pwl.plot(energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2)
# assign covariance for plotting
pwl.parameters.covariance = result_pwl.parameters.covariance
pwl.plot_error(energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2)
ax.set_ylim(1e-13, 1e-11);
Exponential Cut-Off Powerlaw Fit¶
Next we fit an exponential cut-off power law to the data.
[13]:
ecpl = ExpCutoffPowerLawSpectralModel(
index=1.8,
amplitude="2e-12 cm-2 s-1 TeV-1",
reference="1 TeV",
lambda_="0.1 TeV-1",
)
We run the fitter again by passing the flux points and the ecpl model instance:
[14]:
dataset_ecpl = FluxPointsDataset(ecpl, flux_points, likelihood="chi2assym")
fitter = Fit(dataset_ecpl)
result_ecpl = fitter.run()
print(ecpl)
ExpCutoffPowerLawSpectralModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
index 1.869e+00 nan nan nan False
amplitude 2.126e-12 nan cm-2 s-1 TeV-1 nan nan False
reference 1.000e+00 nan TeV nan nan True
lambda_ 1.000e-01 nan TeV-1 nan nan False
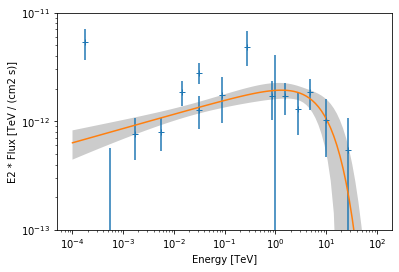
We plot the data and best fit model:
[15]:
ax = flux_points.plot(energy_power=2)
ecpl.plot(energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2)
# assign covariance for plotting
ecpl.parameters.covariance = result_ecpl.parameters.covariance
ecpl.plot_error(energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2)
ax.set_ylim(1e-13, 1e-11)
[15]:
(1e-13, 1e-11)
Log-Parabola Fit¶
Finally we try to fit a log-parabola model:
[16]:
log_parabola = LogParabolaSpectralModel(
alpha=2, amplitude="1e-12 cm-2 s-1 TeV-1", reference="1 TeV", beta=0.1
)
[17]:
dataset_log_parabola = FluxPointsDataset(
log_parabola, flux_points, likelihood="chi2assym"
)
fitter = Fit(dataset_log_parabola)
result_log_parabola = fitter.run()
print(log_parabola)
LogParabolaSpectralModel
Parameters:
name value error unit min max frozen
--------- --------- ----- -------------- --- --- ------
amplitude 1.930e-12 nan cm-2 s-1 TeV-1 nan nan False
reference 1.000e+00 nan TeV nan nan True
alpha 2.151e+00 nan nan nan False
beta 5.259e-02 nan nan nan False
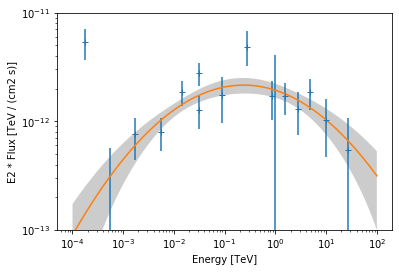
[18]:
ax = flux_points.plot(energy_power=2)
log_parabola.plot(energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2)
# assign covariance for plotting
log_parabola.parameters.covariance = result_log_parabola.parameters.covariance
log_parabola.plot_error(
energy_range=[1e-4, 1e2] * u.TeV, ax=ax, energy_power=2
)
ax.set_ylim(1e-13, 1e-11);
Exercises¶
- Fit a
PowerLaw2SpectralModelandExpCutoffPowerLaw3FGLSpectralModelto the same data. - Fit a
ExpCutoffPowerLawSpectralModelmodel to Vela X (‘HESS J0835-455’) only and check if the best fit values correspond to the values given in the Gammacat catalog
What next?¶
This was an introduction to SED fitting in Gammapy.
- If you would like to learn how to perform a full Poisson maximum likelihood spectral fit, please check out the spectrum analysis tutorial.
- To learn more about other parts of Gammapy (e.g. Fermi-LAT and TeV data analysis), check out the other tutorial notebooks.
- To see what’s available in Gammapy, browse the Gammapy docs or use the full-text search.
- If you have any questions, ask on the mailing list .
[ ]: