EnergyDependentMultiGaussPSF¶
-
class
gammapy.irf.EnergyDependentMultiGaussPSF(energy_lo, energy_hi, theta, sigmas, norms, energy_thresh_lo='0.1 TeV', energy_thresh_hi='100 TeV')[source]¶ Bases:
objectTriple Gauss analytical PSF depending on energy and theta.
To evaluate the PSF call the
to_energy_dependent_table_psforpsf_at_energy_and_thetamethods.Parameters: - energy_lo :
Quantity Lower energy boundary of the energy bin.
- energy_hi :
Quantity Upper energy boundary of the energy bin.
- theta :
Quantity Center values of the theta bins.
- sigmas : list of ‘numpy.ndarray’
Triple Gauss sigma parameters, where every entry is a two dimensional ‘numpy.ndarray’ containing the sigma value for every given energy and theta.
- norms : list of ‘numpy.ndarray’
Triple Gauss norm parameters, where every entry is a two dimensional ‘numpy.ndarray’ containing the norm value for every given energy and theta. Norm corresponds to the value of the Gaussian at theta = 0.
- energy_thresh_lo :
Quantity Lower save energy threshold of the psf.
- energy_thresh_hi :
Quantity Upper save energy threshold of the psf.
Examples
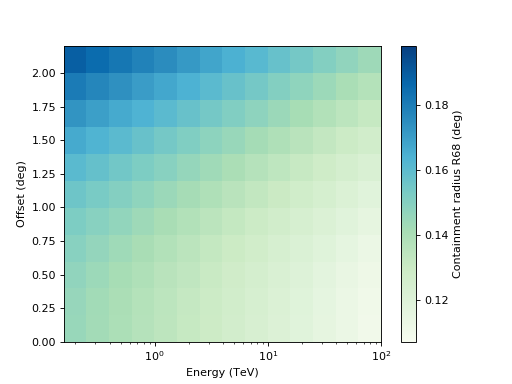
Plot R68 of the PSF vs. theta and energy:
import matplotlib.pyplot as plt from gammapy.irf import EnergyDependentMultiGaussPSF filename = '$GAMMAPY_DATA/tests/unbundled/irfs/psf.fits' psf = EnergyDependentMultiGaussPSF.read(filename, hdu='POINT SPREAD FUNCTION') psf.plot_containment(0.68, show_safe_energy=False) plt.show()
Methods Summary
containment_radius(self, energy, theta[, …])Compute containment for all energy and theta values from_fits(hdu)Create EnergyDependentMultiGaussPSFfrom HDU list.info(self[, fractions, energies, thetas])Print PSF summary info. peek(self[, figsize])Quick-look summary plots. plot_containment(self[, fraction, ax, …])Plot containment image with energy and theta axes. plot_containment_vs_energy(self[, …])Plot containment fraction as a function of energy. psf_at_energy_and_theta(self, energy, theta)Get MultiGauss2Dmodel for given energy and theta.read(filename[, hdu])Create EnergyDependentMultiGaussPSFfrom FITS file.to_energy_dependent_table_psf(self[, theta, …])Convert triple Gaussian PSF ot table PSF. to_fits(self)Convert psf table data to FITS hdu list. to_psf3d(self, rad)Create a PSF3D from an analytical PSF. write(self, filename, \*args, \*\*kwargs)Write PSF to FITS file. Methods Documentation
-
containment_radius(self, energy, theta, fraction=0.68)[source]¶ Compute containment for all energy and theta values
-
classmethod
from_fits(hdu)[source]¶ Create
EnergyDependentMultiGaussPSFfrom HDU list.Parameters: - hdu :
BintableHDU HDU
- hdu :
-
info(self, fractions=[0.68, 0.95], energies=<Quantity [ 1., 10.] TeV>, thetas=<Quantity [0.] deg>)[source]¶ Print PSF summary info.
The containment radius for given fraction, energies and thetas is computed and printed on the command line.
Parameters: Returns: - ss : string
Formatted string containing the summary info.
-
plot_containment(self, fraction=0.68, ax=None, show_safe_energy=False, add_cbar=True, **kwargs)[source]¶ Plot containment image with energy and theta axes.
Parameters: - fraction : float
Containment fraction between 0 and 1.
- add_cbar : bool
Add a colorbar
-
plot_containment_vs_energy(self, fractions=[0.68, 0.95], thetas=<Angle [0., 1.] deg>, ax=None, **kwargs)[source]¶ Plot containment fraction as a function of energy.
-
psf_at_energy_and_theta(self, energy, theta)[source]¶ Get
MultiGauss2Dmodel for given energy and theta.No interpolation is used.
Parameters: Returns: - psf :
MultiGauss2D Multigauss PSF object.
- psf :
-
classmethod
read(filename, hdu='PSF_2D_GAUSS')[source]¶ Create
EnergyDependentMultiGaussPSFfrom FITS file.Parameters: - filename : str
File name
-
to_energy_dependent_table_psf(self, theta=None, rad=None, exposure=None)[source]¶ Convert triple Gaussian PSF ot table PSF.
Parameters: Returns: - tabe_psf :
EnergyDependentTablePSF Instance of
EnergyDependentTablePSF.
- tabe_psf :
-
to_fits(self)[source]¶ Convert psf table data to FITS hdu list.
Returns: - hdu_list :
HDUList PSF in HDU list format.
- hdu_list :
- energy_lo :