This is a fixed-text formatted version of a Jupyter notebook.
- Try online
- You can contribute with your own notebooks in this GitHub repository.
- Source files: cta_sensitivity.ipynb | cta_sensitivity.py
Computation of the CTA sensitivity¶
Introduction¶
This notebook explains how to derive the CTA sensitivity for a point-like IRF at a fixed zenith angle and fixed offset. The significativity is computed for the 1D analysis (On-OFF regions) and the LiMa formula.
We will be using the following Gammapy classes:
Setup¶
As usual, we’ll start with some setup …
In [1]:
%matplotlib inline
import matplotlib.pyplot as plt
In [2]:
from gammapy.irf import CTAPerf
from gammapy.spectrum import SensitivityEstimator
Load IRFs¶
First load the CTA IRFs.
In [3]:
filename = "$GAMMAPY_EXTRA/datasets/cta/perf_prod2/point_like_non_smoothed/South_5h.fits.gz"
irf = CTAPerf.read(filename)
Compute sensitivity¶
Choose a few parameters, then run the sentitivity computation.
In [4]:
sensitivity_estimator = SensitivityEstimator(irf=irf, livetime="5h")
sensitivity_estimator.run()
Results¶
The results are given as an Astropy table.
In [5]:
# Show the results table
sensitivity_estimator.results_table
Out[5]:
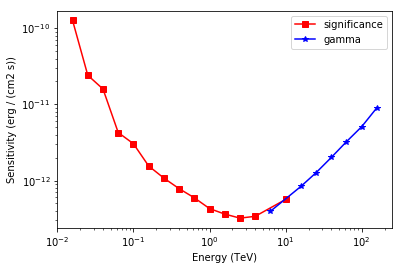
Table length=21
| energy | e2dnde | excess | background | criterion |
|---|---|---|---|---|
| TeV | erg / (cm2 s) | |||
| float32 | float64 | float64 | float32 | str12 |
| 0.0158489 | 1.26569e-10 | 339.143 | 3703.48 | significance |
| 0.0251189 | 2.41235e-11 | 311.106 | 3106.66 | significance |
| 0.0398107 | 1.5914e-11 | 459.213 | 6852.06 | significance |
| 0.0630957 | 4.26714e-12 | 163.204 | 825.794 | significance |
| 0.1 | 3.04454e-12 | 169.361 | 891.645 | significance |
| 0.158489 | 1.55368e-12 | 90.0926 | 236.905 | significance |
| 0.251189 | 1.0771e-12 | 51.5349 | 69.8381 | significance |
| 0.398107 | 7.83236e-13 | 35.6905 | 29.8996 | significance |
| 0.630957 | 5.93807e-13 | 26.4005 | 14.2506 | significance |
| 1 | 4.28759e-13 | 18.1072 | 5.11857 | significance |
| 1.58489 | 3.62852e-13 | 15.6871 | 3.31032 | significance |
| 2.51189 | 3.21257e-13 | 11.7016 | 1.17059 | significance |
| 3.98107 | 3.39152e-13 | 12.0962 | 1.33607 | significance |
| 6.30957 | 3.9511e-13 | 10 | 0.424068 | gamma |
| 10 | 5.65043e-13 | 10.898 | 0.865076 | significance |
| 15.8489 | 8.36566e-13 | 10 | 0.216136 | gamma |
| 25.1189 | 1.26771e-12 | 10 | 0.00979249 | gamma |
| 39.8107 | 2.00893e-12 | 10 | 0.0053102 | gamma |
| 63.0957 | 3.24246e-12 | 10 | 0.00170479 | gamma |
| 100 | 5.10213e-12 | 10 | 0.00101395 | gamma |
| 158.489 | 9.04831e-12 | 10 | 0.00566093 | gamma |
In [6]:
# Save it to file (could use e.g. format of CSV or ECSV or FITS)
# sensitivity_estimator.results_table.write('sensitivity.ecsv', format='ascii.ecsv')
In [7]:
# Plot the sensitivity curve
t = sensitivity_estimator.results_table
is_s = t["criterion"] == "significance"
plt.plot(
t["energy"][is_s],
t["e2dnde"][is_s],
"s-",
color="red",
label="significance",
)
is_g = t["criterion"] == "gamma"
plt.plot(
t["energy"][is_g], t["e2dnde"][is_g], "*-", color="blue", label="gamma"
)
plt.loglog()
plt.xlabel("Energy ({})".format(t["energy"].unit))
plt.ylabel("Sensitivity ({})".format(t["e2dnde"].unit))
plt.legend();
Exercises¶
- Also compute the sensitivity for a 20 hour observation
- Compare how the sensitivity differs between 5 and 20 hours by plotting the ratio as a function of energy.