CountsSpectrum¶
-
class
gammapy.spectrum.CountsSpectrum(energy_lo, energy_hi, data=None, interp_kwargs=None)[source]¶ Bases:
objectGeneric counts spectrum.
Parameters: Examples
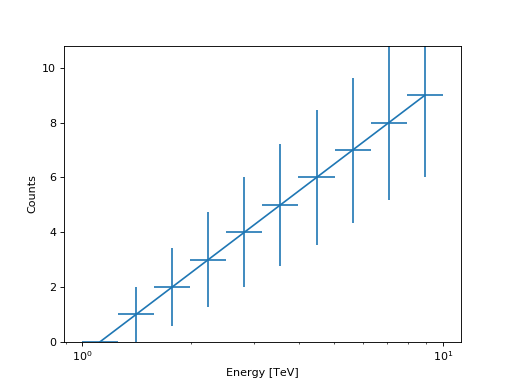
from gammapy.spectrum import CountsSpectrum import numpy as np import astropy.units as u ebounds = np.logspace(0,1,11) * u.TeV data = np.arange(10) spec = CountsSpectrum( energy_lo=ebounds[:-1], energy_hi=ebounds[1:], data=data, ) spec.plot(show_poisson_errors=True)
Attributes Summary
default_interp_kwargsDefault interpolation kwargs energytotal_countsTotal number of counts Methods Summary
copy(self)A deep copy of self. energy_mask(self[, emin, emax])Create a mask for a given energy range. fill(self, events)Fill with list of events. from_hdulist(hdulist[, hdu1, hdu2])Read OGIP format hdulist peek(self[, figsize])Quick-look summary plots. plot(self[, ax, energy_unit, …])Plot as data points. plot_hist(self[, ax, energy_unit, show_energy])Plot as histogram. read(filename[, hdu1, hdu2])Read from file. rebin(self, parameter)Rebin. to_hdulist(self[, use_sherpa])Convert to HDUList.to_table(self)Convert to Table.write(self, filename[, use_sherpa])Write to file. Attributes Documentation
-
default_interp_kwargs= {'bounds_error': False, 'method': 'nearest'}¶ Default interpolation kwargs
-
energy¶
-
total_counts¶ Total number of counts
Methods Documentation
-
energy_mask(self, emin=None, emax=None)[source]¶ Create a mask for a given energy range.
Parameters: - emin, emax :
Quantity Energy range
- emin, emax :
-
fill(self, events)[source]¶ Fill with list of events.
TODO: Move to
NDDataArrayParameters: - events :
Quantity,gammapy.data.EventList, List of event energies
- events :
-
plot(self, ax=None, energy_unit='TeV', show_poisson_errors=False, show_energy=None, **kwargs)[source]¶ Plot as data points.
kwargs are forwarded to
errorbarParameters: Returns: - ax:
axis Axis instance used for the plot
- ax:
-
plot_hist(self, ax=None, energy_unit='TeV', show_energy=None, **kwargs)[source]¶ Plot as histogram.
kwargs are forwarded to
histParameters:
-
rebin(self, parameter)[source]¶ Rebin.
Parameters: - parameter : int
Number of bins to merge
Returns: - rebinned_spectrum :
CountsSpectrum Rebinned spectrum
-