This is a fixed-text formatted version of a Jupyter notebook
You may download all the notebooks in the documentation as a tar file.
Source files: modeling_2D.ipynb | modeling_2D.py
2D map fitting¶
Prerequisites:¶
To understand how a generel modelling and fiiting works in gammapy, please refer to the analysis_3d tutorial
Context:¶
We often want the determine the position and morphology of an object. To do so, we don’t necessarily have to resort to a full 3D fitting but can perform a simple image fitting, in particular, in an energy range where the PSF does not vary strongly, or if we want to explore a possible energy dependence of the morphology.
Objective:¶
To localize a source and/or constrain its morphology.
Proposed approach:¶
The first step here, as in most analysis with DL3 data, is to create reduced datasets. For this, we will use the Analysis class to create a single set of stacked maps with a single bin in energy (thus, an image which behaves as a cube). This, we will then model with a spatial model of our choice, while keeping the spectral model fixed to an integrated power law.
Setup¶
As usual, we’ll start with some general imports…
[1]:
%matplotlib inline
import astropy.units as u
from astropy.coordinates import SkyCoord
from astropy.time import Time
import logging
log = logging.getLogger(__name__)
Now let’s import gammapy specific classes and functions
[2]:
from gammapy.analysis import Analysis, AnalysisConfig
Creating the config file¶
Now, we create a config file for out analysis. You may load this from disc if you have a pre-defined config file.
Here, we use 3 simulated CTA runs of the galactic center.
[3]:
config = AnalysisConfig()
# Selecting the observations
config.observations.datastore = "$GAMMAPY_DATA/cta-1dc/index/gps/"
config.observations.obs_ids = [110380, 111140, 111159]
Technically, gammapy implements 2D analysis as a special case of 3D analysis (one one bin in energy). So, we must specify the type of analysis as 3D, and define the geometry of the analysis.
[4]:
config.datasets.type = "3d"
config.datasets.geom.wcs.skydir = {
"lon": "0 deg",
"lat": "0 deg",
"frame": "galactic",
} # The WCS geometry - centered on the galactic center
config.datasets.geom.wcs.width = {"width": "8 deg", "height": "6 deg"}
config.datasets.geom.wcs.binsize = "0.02 deg"
# The FoV radius to use for cutouts
config.datasets.geom.selection.offset_max = 2.5 * u.deg
config.datasets.safe_mask.methods = ["offset-max"]
config.datasets.safe_mask.parameters = {"offset_max": 2.5 * u.deg}
config.datasets.background.method = "fov_background"
config.fit.fit_range = {"min": "0.1 TeV", "max": "30.0 TeV"}
# We now fix the energy axis for the counts map - (the reconstructed energy binning)
config.datasets.geom.axes.energy.min = "0.1 TeV"
config.datasets.geom.axes.energy.max = "10 TeV"
config.datasets.geom.axes.energy.nbins = 1
config.datasets.geom.wcs.binsize_irf = 0.2 * u.deg
[5]:
print(config)
AnalysisConfig
general:
log: {level: info, filename: null, filemode: null, format: null, datefmt: null}
outdir: .
observations:
datastore: $GAMMAPY_DATA/cta-1dc/index/gps
obs_ids: [110380, 111140, 111159]
obs_file: null
obs_cone: {frame: null, lon: null, lat: null, radius: null}
obs_time: {start: null, stop: null}
required_irf: [aeff, edisp, psf, bkg]
datasets:
type: 3d
stack: true
geom:
wcs:
skydir: {frame: galactic, lon: 0.0 deg, lat: 0.0 deg}
binsize: 0.02 deg
width: {width: 8.0 deg, height: 6.0 deg}
binsize_irf: 0.2 deg
selection: {offset_max: 2.5 deg}
axes:
energy: {min: 0.1 TeV, max: 10.0 TeV, nbins: 1}
energy_true: {min: 0.5 TeV, max: 20.0 TeV, nbins: 16}
map_selection: [counts, exposure, background, psf, edisp]
background:
method: fov_background
exclusion: null
parameters: {}
safe_mask:
methods: [offset-max]
parameters: {offset_max: 2.5 deg}
on_region: {frame: null, lon: null, lat: null, radius: null}
containment_correction: true
fit:
fit_range: {min: 0.1 TeV, max: 30.0 TeV}
flux_points:
energy: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
excess_map:
correlation_radius: 0.1 deg
parameters: {}
energy_edges: {min: null, max: null, nbins: null}
light_curve:
time_intervals: {start: null, stop: null}
energy_edges: {min: null, max: null, nbins: null}
source: source
parameters: {selection_optional: all}
Getting the reduced dataset¶
We now use the config file and create a single MapDataset containing counts, background, exposure, psf and edisp maps.
[6]:
%%time
analysis = Analysis(config)
analysis.get_observations()
analysis.get_datasets()
Setting logging config: {'level': 'INFO', 'filename': None, 'filemode': None, 'format': None, 'datefmt': None}
Fetching observations.
No HDU found matching: OBS_ID = 110380, HDU_TYPE = rad_max, HDU_CLASS = None
No HDU found matching: OBS_ID = 111140, HDU_TYPE = rad_max, HDU_CLASS = None
No HDU found matching: OBS_ID = 111159, HDU_TYPE = rad_max, HDU_CLASS = None
Number of selected observations: 3
Creating reference dataset and makers.
Creating the background Maker.
Start the data reduction loop.
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
Invalid unit found in background table! Assuming (s-1 MeV-1 sr-1)
CPU times: user 3.19 s, sys: 444 ms, total: 3.63 s
Wall time: 3.63 s
[7]:
print(analysis.datasets["stacked"])
MapDataset
----------
Name : stacked
Total counts : 85625
Total background counts : 85624.99
Total excess counts : 0.01
Predicted counts : 85625.00
Predicted background counts : 85624.99
Predicted excess counts : nan
Exposure min : 8.46e+08 m2 s
Exposure max : 2.14e+10 m2 s
Number of total bins : 120000
Number of fit bins : 96602
Fit statistic type : cash
Fit statistic value (-2 log(L)) : nan
Number of models : 0
Number of parameters : 0
Number of free parameters : 0
The counts and background maps have only one bin in reconstructed energy. The exposure and IRF maps are in true energy, and hence, have a different binning based upon the binning of the IRFs. We need not bother about them presently.
[8]:
analysis.datasets["stacked"].counts
[8]:
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (400, 300, 1)
ndim : 3
unit :
dtype : float32
[9]:
analysis.datasets["stacked"].background
[9]:
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy']
shape : (400, 300, 1)
ndim : 3
unit :
dtype : float32
[10]:
analysis.datasets["stacked"].exposure
[10]:
WcsNDMap
geom : WcsGeom
axes : ['lon', 'lat', 'energy_true']
shape : (400, 300, 16)
ndim : 3
unit : m2 s
dtype : float32
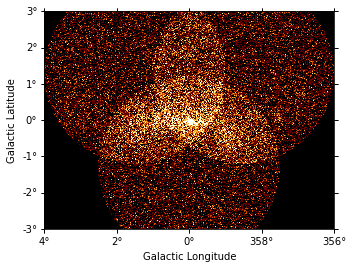
We can have a quick look of these maps in the following way:
[11]:
analysis.datasets["stacked"].counts.reduce_over_axes().plot(vmax=5)
/Users/adonath/software/mambaforge/envs/gammapy-dev/lib/python3.9/site-packages/astropy/visualization/wcsaxes/core.py:211: MatplotlibDeprecationWarning: Passing parameters norm and vmin/vmax simultaneously is deprecated since 3.3 and will become an error two minor releases later. Please pass vmin/vmax directly to the norm when creating it.
return super().imshow(X, *args, origin=origin, **kwargs)
[11]:
<WCSAxesSubplot:xlabel='Galactic Longitude', ylabel='Galactic Latitude'>
Modelling¶
Now, we define a model to be fitted to the dataset. The important thing to note here is the dummy spectral model - an integrated powerlaw with only free normalisation. Here, we use its YAML definition to load it:
[12]:
model_config = """
components:
- name: GC-1
type: SkyModel
spatial:
type: PointSpatialModel
frame: galactic
parameters:
- name: lon_0
value: 0.02
unit: deg
- name: lat_0
value: 0.01
unit: deg
spectral:
type: PowerLaw2SpectralModel
parameters:
- name: amplitude
value: 1.0e-12
unit: cm-2 s-1
- name: index
value: 2.0
unit: ''
frozen: true
- name: emin
value: 0.1
unit: TeV
frozen: true
- name: emax
value: 10.0
unit: TeV
frozen: true
"""
[13]:
analysis.set_models(model_config)
Reading model.
Models
Component 0: SkyModel
Name : GC-1
Datasets names : None
Spectral model type : PowerLaw2SpectralModel
Spatial model type : PointSpatialModel
Temporal model type :
Parameters:
amplitude : 1.00e-12 +/- 0.0e+00 1 / (cm2 s)
index (frozen) : 2.000
emin (frozen) : 0.100 TeV
emax (frozen) : 10.000 TeV
lon_0 : 0.020 +/- 0.00 deg
lat_0 : 0.010 +/- 0.00 deg
Component 1: FoVBackgroundModel
Name : stacked-bkg
Datasets names : ['stacked']
Spectral model type : PowerLawNormSpectralModel
Parameters:
norm : 1.000 +/- 0.00
tilt (frozen) : 0.000
reference (frozen) : 1.000 TeV
We will freeze the parameters of the background
[14]:
analysis.datasets["stacked"].background_model.parameters["tilt"].frozen = True
[15]:
# To run the fit
analysis.run_fit()
Fitting datasets.
OptimizeResult
backend : minuit
method : migrad
success : True
message : Optimization terminated successfully.
nfev : 175
total stat : 170089.04
OptimizeResult
backend : minuit
method : migrad
success : True
message : Optimization terminated successfully.
nfev : 175
total stat : 170089.04
[16]:
# To see the best fit values along with the errors
analysis.models.to_parameters_table()
[16]:
| model | type | name | value | unit | error | min | max | frozen | link |
|---|---|---|---|---|---|---|---|---|---|
| str11 | str8 | str9 | float64 | str8 | float64 | float64 | float64 | bool | str1 |
| GC-1 | spectral | amplitude | 4.1790e-11 | cm-2 s-1 | 2.230e-12 | nan | nan | False | |
| GC-1 | spectral | index | 2.0000e+00 | 0.000e+00 | nan | nan | True | ||
| GC-1 | spectral | emin | 1.0000e-01 | TeV | 0.000e+00 | nan | nan | True | |
| GC-1 | spectral | emax | 1.0000e+01 | TeV | 0.000e+00 | nan | nan | True | |
| GC-1 | spatial | lon_0 | -5.4771e-02 | deg | 1.992e-03 | nan | nan | False | |
| GC-1 | spatial | lat_0 | -5.3620e-02 | deg | 1.981e-03 | -9.000e+01 | 9.000e+01 | False | |
| stacked-bkg | spectral | norm | 9.9439e-01 | 3.411e-03 | nan | nan | False | ||
| stacked-bkg | spectral | tilt | 0.0000e+00 | 0.000e+00 | nan | nan | True | ||
| stacked-bkg | spectral | reference | 1.0000e+00 | TeV | 0.000e+00 | nan | nan | True |
[ ]:
