Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Morphological energy dependence estimation#
Learn how to test for energy-dependent morphology in your dataset.
Prerequisites#
Knowledge on data reduction and datasets used in Gammapy, for example see the H.E.S.S. with Gammapy and Ring background map tutorials.
Context#
This tutorial introduces a method to investigate the potential of energy-dependent morphology from spatial maps. It is possible for gamma-ray sources to exhibit energy-dependent morphology, in which the spatial morphology of the gamma rays varies across different energy bands. This is plausible for different source types, including pulsar wind nebulae (PWNe) and supernova remnants. HESS J1825−137 is a well-known example of a PWNe which shows a clear energy-dependent gamma-ray morphology (see Aharonian et al., 2006, H.E.S.S. Collaboration et al., 2019 and Principe et al., 2020.)
Many different techniques of measuring this energy-dependence have been utilised over the years. The method shown here is to perform a morphological fit of extension and position in various energy slices and compare this with a global morphology fit.
Objective: Perform an energy-dependent morphology study of a mock source.
Tutorial overview#
This tutorial consists of two main steps.
Firstly, the user defines the initial SkyModel based on previous investigations
and selects the energy bands of interest to test for energy dependence. The null hypothesis is defined as
only the background component being free (norm). The alternative hypothesis introduces the source model.
The results of this first step show the significance of the source above the background in each energy band.
The second step is to quantify any energy-dependent morphology. The null hypothesis is determined by performing a joint fit of the parameters. In the alternative hypothesis, the free parameters of the model are fit individually within each energy band.
Setup#
from astropy import units as u
from astropy.coordinates import SkyCoord
from astropy.table import Table
import matplotlib.pyplot as plt
from IPython.display import display
from gammapy.datasets import Datasets, MapDataset
from gammapy.estimators import EnergyDependentMorphologyEstimator
from gammapy.estimators.energydependentmorphology import weighted_chi2_parameter
from gammapy.maps import Map
from gammapy.modeling.models import (
GaussianSpatialModel,
PowerLawSpectralModel,
SkyModel,
)
from gammapy.stats.utils import ts_to_sigma
from gammapy.utils.check import check_tutorials_setup
Check setup#
check_tutorials_setup()
System:
python_executable : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/bin/python
python_version : 3.9.22
machine : x86_64
system : Linux
Gammapy package:
version : 2.0.dev1224+g83b25692c
path : /home/runner/work/gammapy-docs/gammapy-docs/gammapy/.tox/build_docs/lib/python3.9/site-packages/gammapy
Other packages:
numpy : 1.26.4
scipy : 1.13.1
astropy : 6.0.1
regions : 0.8
click : 8.1.8
yaml : 6.0.2
IPython : 8.18.1
jupyterlab : not installed
matplotlib : 3.9.4
pandas : not installed
healpy : 1.17.3
iminuit : 2.31.1
sherpa : 4.16.1
naima : 0.10.0
emcee : 3.1.6
corner : 2.2.3
ray : 2.46.0
Gammapy environment variables:
GAMMAPY_DATA : /home/runner/work/gammapy-docs/gammapy-docs/gammapy-datasets/dev
Obtain the data to use#
Utilise the pre-defined dataset within $GAMMAPY_DATA.
P.S.: do not forget to set up your environment variable $GAMMAPY_DATA
to your local directory.
stacked_dataset = MapDataset.read(
"$GAMMAPY_DATA/estimators/mock_DL4/dataset_energy_dependent.fits.gz"
)
datasets = Datasets([stacked_dataset])
Define the energy edges of interest. These will be utilised to investigate the potential of energy-dependent morphology in the dataset.
energy_edges = [1, 3, 5, 20] * u.TeV
Define the spectral and spatial models of interest. We utilise
a PowerLawSpectralModel and a
GaussianSpatialModel to test the energy-dependent
morphology component in each energy band. A standard 3D fit (see the
3D detailed analysis tutorial)
is performed, then the best fit model is utilised here for the initial parameters
in each model.
source_position = SkyCoord(5.58, 0.2, unit="deg", frame="galactic")
spectral_model = PowerLawSpectralModel(
index=2.5, amplitude=9.8e-12 * u.Unit("cm-2 s-1 TeV-1"), reference=1.0 * u.TeV
)
spatial_model = GaussianSpatialModel(
lon_0=source_position.l,
lat_0=source_position.b,
frame="galactic",
sigma=0.2 * u.deg,
)
# Limit the search for the position on the spatial model
spatial_model.lon_0.min = source_position.galactic.l.deg - 0.8
spatial_model.lon_0.max = source_position.galactic.l.deg + 0.8
spatial_model.lat_0.min = source_position.galactic.b.deg - 0.8
spatial_model.lat_0.max = source_position.galactic.b.deg + 0.8
model = SkyModel(spatial_model=spatial_model, spectral_model=spectral_model, name="src")
Run Estimator#
We can now run the energy-dependent estimation tool and explore the results.
Let’s start with the initial hypothesis, in which the source is introduced to compare with the background. We specify which parameters we wish to use for testing the energy dependence. To test for the energy dependence, it is recommended to keep the position and extension parameters free. This allows them to be used for fitting the spatial model in each energy band.
model.spatial_model.lon_0.frozen = False
model.spatial_model.lat_0.frozen = False
model.spatial_model.sigma.frozen = False
model.spectral_model.amplitude.frozen = False
model.spectral_model.index.frozen = True
datasets.models = model
estimator = EnergyDependentMorphologyEstimator(energy_edges=energy_edges, source="src")
Show the results tables#
The results of the source signal above the background in each energy bin#
Firstly, the estimator is run to produce the results. The table here shows the ∆(TS) value, the number of degrees of freedom (df) and the significance (σ) in each energy bin. The significance values here show that each energy band has significant signal above the background.
results = estimator.run(datasets)
table_bkg_src = Table(results["src_above_bkg"])
display(table_bkg_src)
Emin Emax delta_ts df significance
TeV TeV
---- ---- ----------------- --- ------------------
1.0 3.0 998.0521468441366 4 31.277522361013748
3.0 5.0 712.8736082200157 4 26.34613054105945
5.0 20.0 290.414054187946 4 16.564139576922415
The results for testing energy dependence#
Next, the results of the energy-dependent estimator are shown.
The table shows the various free parameters for the joint fit for
ts = results["energy_dependence"]["delta_ts"]
df = results["energy_dependence"]["df"]
sigma = ts_to_sigma(ts, df=df)
print(f"The delta_ts for the energy-dependent study: {ts:.3f}.")
print(f"Converting this to a significance gives: {sigma:.3f} \u03C3")
results_table = Table(results["energy_dependence"]["result"])
display(results_table)
The delta_ts for the energy-dependent study: 76.825.
Converting this to a significance gives: 7.678 σ
Hypothesis Emin Emax ... sigma sigma_err
TeV TeV ... deg deg
---------- ---- ---- ... ------------------- --------------------
H0 1.0 20.0 ... 0.21525839967018867 0.005911103564621735
H1 1.0 3.0 ... 0.2568710837134529 0.009428253484623464
H1 3.0 5.0 ... 0.19736017641361556 0.008166963876141447
H1 5.0 20.0 ... 0.13499879586502125 0.00889194478934663
The chi-squared value for each parameter of interest#
We can also utilise the weighted_chi2_parameter
function for each parameter.
The weighted chi-squared significance for the sigma, lat_0 and lon_0 values.
parameter chi2 df significance
--------- ------------------ --- ------------------
sigma 88.6610048102039 2 9.151955134028793
lat_0 4.691492300600545 2 1.6656873021399468
lon_0 1.3055487448271073 2 0.6424216679053606
Note: The chi-squared parameter does not include potential correlation between the parameters, so it should be used cautiously.
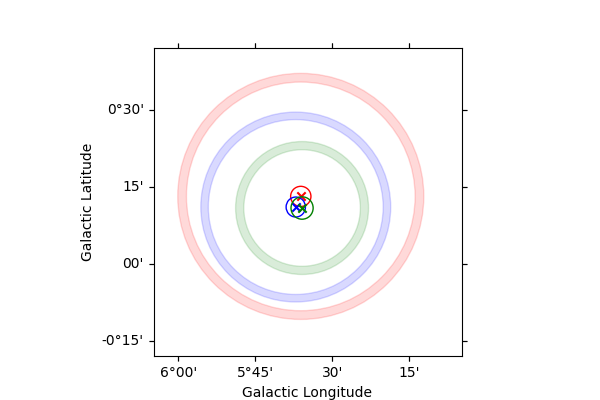
Plotting the results#
empty_map = Map.create(
skydir=spatial_model.position, frame=spatial_model.frame, width=1, binsz=0.02
)
colors = ["red", "blue", "green", "magenta"]
fig = plt.figure(figsize=(6, 4))
ax = empty_map.plot()
lat_0 = results["energy_dependence"]["result"]["lat_0"][1:]
lat_0_err = results["energy_dependence"]["result"]["lat_0_err"][1:]
lon_0 = results["energy_dependence"]["result"]["lon_0"][1:]
lon_0_err = results["energy_dependence"]["result"]["lon_0_err"][1:]
sigma = results["energy_dependence"]["result"]["sigma"][1:]
sigma_err = results["energy_dependence"]["result"]["sigma_err"][1:]
for i in range(len(lat_0)):
model_plot = GaussianSpatialModel(
lat_0=lat_0[i], lon_0=lon_0[i], sigma=sigma[i], frame=spatial_model.frame
)
model_plot.lat_0.error = lat_0_err[i]
model_plot.lon_0.error = lon_0_err[i]
model_plot.sigma.error = sigma_err[i]
model_plot.plot_error(
ax=ax,
which="all",
kwargs_extension={"facecolor": colors[i], "edgecolor": colors[i]},
kwargs_position={"color": colors[i]},
)
plt.show()
Total running time of the script: (0 minutes 19.408 seconds)
